The relation between recombination rate and patterns of molecular evolution and variation in Drosophila melanogaster
- PMID: 24489114
- PMCID: PMC3969569
- DOI: 10.1093/molbev/msu056
The relation between recombination rate and patterns of molecular evolution and variation in Drosophila melanogaster
Abstract
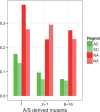
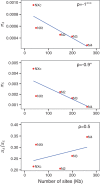
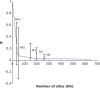
Genetic recombination associated with sexual reproduction increases the efficiency of natural selection by reducing the strength of Hill-Robertson interference. Such interference can be caused either by selective sweeps of positively selected alleles or by background selection (BGS) against deleterious mutations. Its consequences can be studied by comparing patterns of molecular evolution and variation in genomic regions with different rates of crossing over. We carried out a comprehensive study of the benefits of recombination in Drosophila melanogaster, both by contrasting five independent genomic regions that lack crossing over with the rest of the genome and by comparing regions with different rates of crossing over, using data on DNA sequence polymorphisms from an African population that is geographically close to the putatively ancestral population for the species, and on sequence divergence from a related species. We observed reductions in sequence diversity in noncrossover (NC) regions that are inconsistent with the effects of hard selective sweeps in the absence of recombination. Overall, the observed patterns suggest that the recombination rate experienced by a gene is positively related to an increase in the efficiency of both positive and purifying selection. The results are consistent with a BGS model with interference among selected sites in NC regions, and joint effects of BGS, selective sweeps, and a past population expansion on variability in regions of the genome that experience crossing over. In such crossover regions, the X chromosome exhibits a higher rate of adaptive protein sequence evolution than the autosomes, implying a Faster-X effect.
Keywords: Drosophila melanogaster; Hill–Robertson interference; background selection; crossing over; heterochromatin; recombination; selective sweeps.
Figures

References
-
- Andolfatto P. Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol Biol Evol. 2001;18:279–290. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

