Semantic particularity measure for functional characterization of gene sets using gene ontology
- PMID: 24489737
- PMCID: PMC3904913
- DOI: 10.1371/journal.pone.0086525
Semantic particularity measure for functional characterization of gene sets using gene ontology
Abstract
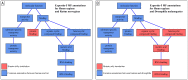
Background: Genetic and genomic data analyses are outputting large sets of genes. Functional comparison of these gene sets is a key part of the analysis, as it identifies their shared functions, and the functions that distinguish each set. The Gene Ontology (GO) initiative provides a unified reference for analyzing the genes molecular functions, biological processes and cellular components. Numerous semantic similarity measures have been developed to systematically quantify the weight of the GO terms shared by two genes. We studied how gene set comparisons can be improved by considering gene set particularity in addition to gene set similarity.
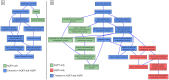
Results: We propose a new approach to compute gene set particularities based on the information conveyed by GO terms. A GO term informativeness can be computed using either its information content based on the term frequency in a corpus, or a function of the term's distance to the root. We defined the semantic particularity of a set of GO terms Sg1 compared to another set of GO terms Sg2. We combined our particularity measure with a similarity measure to compare gene sets. We demonstrated that the combination of semantic similarity and semantic particularity measures was able to identify genes with particular functions from among similar genes. This differentiation was not recognized using only a semantic similarity measure.
Conclusion: Semantic particularity should be used in conjunction with semantic similarity to perform functional analysis of GO-annotated gene sets. The principle is generalizable to other ontologies.
Conflict of interest statement
Figures
Similar articles
-
Optimal Threshold Determination for Interpreting Semantic Similarity and Particularity: Application to the Comparison of Gene Sets and Metabolic Pathways Using GO and ChEBI.PLoS One. 2015 Jul 31;10(7):e0133579. doi: 10.1371/journal.pone.0133579. eCollection 2015. PLoS One. 2015. PMID: 26230274 Free PMC article.
-
Interspecies gene function prediction using semantic similarity.BMC Syst Biol. 2016 Dec 23;10(Suppl 4):121. doi: 10.1186/s12918-016-0361-5. BMC Syst Biol. 2016. PMID: 28155711 Free PMC article.
-
Measure the Semantic Similarity of GO Terms Using Aggregate Information Content.IEEE/ACM Trans Comput Biol Bioinform. 2014 May-Jun;11(3):468-76. doi: 10.1109/TCBB.2013.176. IEEE/ACM Trans Comput Biol Bioinform. 2014. PMID: 26356015
-
TopoICSim: a new semantic similarity measure based on gene ontology.BMC Bioinformatics. 2016 Jul 29;17(1):296. doi: 10.1186/s12859-016-1160-0. BMC Bioinformatics. 2016. PMID: 27473391 Free PMC article.
-
From ontology to semantic similarity: calculation of ontology-based semantic similarity.ScientificWorldJournal. 2013;2013:793091. doi: 10.1155/2013/793091. Epub 2013 Feb 28. ScientificWorldJournal. 2013. PMID: 23533360 Free PMC article. Review.
Cited by
-
Representing virus-host interactions and other multi-organism processes in the Gene Ontology.BMC Microbiol. 2015 Jul 28;15:146. doi: 10.1186/s12866-015-0481-x. BMC Microbiol. 2015. PMID: 26215368 Free PMC article.
-
Co-complex protein membership evaluation using Maximum Entropy on GO ontology and InterPro annotation.Bioinformatics. 2018 Jun 1;34(11):1884-1892. doi: 10.1093/bioinformatics/btx803. Bioinformatics. 2018. PMID: 29390084 Free PMC article.
-
Integrating Information in Biological Ontologies and Molecular Networks to Infer Novel Terms.Sci Rep. 2016 Dec 15;6:39237. doi: 10.1038/srep39237. Sci Rep. 2016. PMID: 27976738 Free PMC article.
-
Identification of Chemical Toxicity Using Ontology Information of Chemicals.Comput Math Methods Med. 2015;2015:246374. doi: 10.1155/2015/246374. Epub 2015 Oct 5. Comput Math Methods Med. 2015. PMID: 26508991 Free PMC article.
-
Optimal Threshold Determination for Interpreting Semantic Similarity and Particularity: Application to the Comparison of Gene Sets and Metabolic Pathways Using GO and ChEBI.PLoS One. 2015 Jul 31;10(7):e0133579. doi: 10.1371/journal.pone.0133579. eCollection 2015. PLoS One. 2015. PMID: 26230274 Free PMC article.
References
-
- Grossmann S, Bauer S, Robinson PN, Vingron M (2007) Improved detection of overrepresentation of gene-ontology annotations with parent child analysis. Bioinformatics 23: 3024–31. - PubMed
-
- Klie S, Mutwil M, Persson S, Nikoloski Z (2012) Inferring gene functions through dissection of relevance networks: interleaving the intra- and inter-species views. Mol Biosyst 8: 2233–41. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

