Comprehensive analysis to improve the validation rate for single nucleotide variants detected by next-generation sequencing
- PMID: 24489763
- PMCID: PMC3906084
- DOI: 10.1371/journal.pone.0086664
Comprehensive analysis to improve the validation rate for single nucleotide variants detected by next-generation sequencing
Abstract
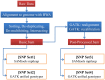
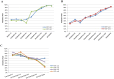
Next-generation sequencing (NGS) has enabled the high-throughput discovery of germline and somatic mutations. However, NGS-based variant detection is still prone to errors, resulting in inaccurate variant calls. Here, we categorized the variants detected by NGS according to total read depth (TD) and SNP quality (SNPQ), and performed Sanger sequencing with 348 selected non-synonymous single nucleotide variants (SNVs) for validation. Using the SAMtools and GATK algorithms, the validation rate was positively correlated with SNPQ but showed no correlation with TD. In addition, common variants called by both programs had a higher validation rate than caller-specific variants. We further examined several parameters to improve the validation rate, and found that strand bias (SB) was a key parameter. SB in NGS data showed a strong difference between the variants passing validation and those that failed validation, showing a validation rate of more than 92% (filtering cutoff value: alternate allele forward [AF] ≥ 20 and AF<80 in SAMtools, SB<-10 in GATK). Moreover, the validation rate increased significantly (up to 97-99%) when the variant was filtered together with the suggested values of mapping quality (MQ), SNPQ and SB. This detailed and systematic study provides comprehensive recommendations for improving validation rates, saving time and lowering cost in NGS analyses.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

