NMR spectroscopic and bioinformatic analyses of the LTBP1 C-terminus reveal a highly dynamic domain organisation
- PMID: 24489852
- PMCID: PMC3906135
- DOI: 10.1371/journal.pone.0087125
NMR spectroscopic and bioinformatic analyses of the LTBP1 C-terminus reveal a highly dynamic domain organisation
Abstract
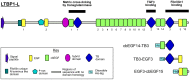
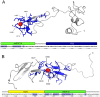
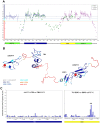
Proteins from the LTBP/fibrillin family perform key structural and functional roles in connective tissues. LTBP1 forms the large latent complex with TGFβ and its propeptide LAP, and sequesters the latent growth factor to the extracellular matrix. Bioinformatics studies suggest the main structural features of the LTBP1 C-terminus are conserved through evolution. NMR studies were carried out on three overlapping C-terminal fragments of LTBP1, comprising four domains with characterised homologues, cbEGF14, TB3, EGF3 and cbEGF15, and three regions with no homology to known structures. The NMR data reveal that the four domains adopt canonical folds, but largely lack the interdomain interactions observed with homologous fibrillin domains; the exception is the EGF3-cbEGF15 domain pair which has a well-defined interdomain interface. (15)N relaxation studies further demonstrate that the three interdomain regions act as flexible linkers, allowing a wide range of motion between the well-structured domains. This work is consistent with the LTBP1 C-terminus adopting a flexible "knotted rope" structure, which may facilitate cell matrix interactions, and the accessibility to proteases or other factors that could contribute to TGFβ activation.
Conflict of interest statement
Figures



Similar articles
-
The N-Terminal Region of Fibrillin-1 Mediates a Bipartite Interaction with LTBP1.Structure. 2017 Aug 1;25(8):1208-1221.e5. doi: 10.1016/j.str.2017.06.003. Epub 2017 Jun 29. Structure. 2017. PMID: 28669633 Free PMC article.
-
Backbone ¹H, ¹³C and ¹⁵N resonance assignment of the C-terminal EGF-cbEGF pair of LTBP1 and flanking residues.Biomol NMR Assign. 2014 Apr;8(1):159-63. doi: 10.1007/s12104-013-9474-6. Epub 2013 Mar 14. Biomol NMR Assign. 2014. PMID: 23494870
-
Solution structure of the third TB domain from LTBP1 provides insight into assembly of the large latent complex that sequesters latent TGF-beta.J Mol Biol. 2003 Nov 21;334(2):281-91. doi: 10.1016/j.jmb.2003.09.053. J Mol Biol. 2003. PMID: 14607119
-
Latent transforming growth factor-beta binding proteins (LTBPs)--structural extracellular matrix proteins for targeting TGF-beta action.Cytokine Growth Factor Rev. 1999 Jun;10(2):99-117. doi: 10.1016/s1359-6101(99)00010-6. Cytokine Growth Factor Rev. 1999. PMID: 10743502 Review.
-
TB domain proteins: evolutionary insights into the multifaceted roles of fibrillins and LTBPs.Biochem J. 2011 Jan 15;433(2):263-76. doi: 10.1042/BJ20101320. Biochem J. 2011. PMID: 21175431 Review.
Cited by
-
Latent TGF-beta binding protein-1 plays an important role in craniofacial development.J Appl Oral Sci. 2020 Nov 4;28:e20200262. doi: 10.1590/1678-7757-2020-0262. eCollection 2020. J Appl Oral Sci. 2020. PMID: 35320333 Free PMC article.
-
Independent multimerization of Latent TGFβ Binding Protein-1 stabilized by cross-linking and enhanced by heparan sulfate.Sci Rep. 2016 Sep 28;6:34347. doi: 10.1038/srep34347. Sci Rep. 2016. PMID: 27677855 Free PMC article.
-
Latent TGFβ complexes are transglutaminase cross-linked to fibrillin to facilitate TGFβ activation.Matrix Biol. 2022 Mar;107:24-39. doi: 10.1016/j.matbio.2022.01.005. Epub 2022 Feb 3. Matrix Biol. 2022. PMID: 35122964 Free PMC article.
-
Regulation of the Bioavailability of TGF-β and TGF-β-Related Proteins.Cold Spring Harb Perspect Biol. 2016 Jun 1;8(6):a021907. doi: 10.1101/cshperspect.a021907. Cold Spring Harb Perspect Biol. 2016. PMID: 27252363 Free PMC article. Review.
-
Transforming growth factor β latency: A mechanism of cytokine storage and signalling regulation in liver homeostasis and disease.JHEP Rep. 2021 Nov 18;4(2):100397. doi: 10.1016/j.jhepr.2021.100397. eCollection 2022 Feb. JHEP Rep. 2021. PMID: 35059619 Free PMC article. Review.
References
-
- Robertson I, Jensen S, Handford P (2011) TB domain proteins: evolutionary insights into the multifaceted roles of fibrillins and LTBPs. Biochem J 433: 263–276. - PubMed
-
- Massague J (1990) The transforming growth factor-beta family. Annu Rev Cell Biol 6: 597–641. - PubMed
-
- Hyytiainen M, Penttinen C, Keski-Oja J (2004) Latent TGF-beta binding proteins: extracellular matrix association and roles in TGF-beta activation. Crit Rev Clin Lab Sci 41: 233–264. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

