Genetic distinctiveness of the Herdwick sheep breed and two other locally adapted hill breeds of the UK
- PMID: 24489968
- PMCID: PMC3906253
- DOI: 10.1371/journal.pone.0087823
Genetic distinctiveness of the Herdwick sheep breed and two other locally adapted hill breeds of the UK
Abstract
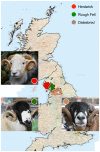
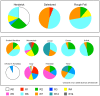
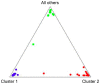
There is considerable interest in locally adapted breeds of livestock as reservoirs of genetic diversity that may provide important fitness traits for future use in agriculture. In marginal areas, these animals contribute to food security and extract value from land unsuitable for other systems of farming. In England, close to 50% of the national sheep flock is farmed on grassland designated as disadvantaged areas for agricultural production. Many of these areas are in the uplands, where some native breeds of sheep continue to be commercially farmed only in highly localised geographical regions to which they are adapted. This study focuses on three of these breeds, selected for their adaptation to near identical environments and their geographical concentration in regions close to one another. Our objective has been to use retrotyping, microsatellites and single nucleotide polymorphisms to explore the origins of the breeds and whether, despite their similar adaptations and proximity, they are genetically distinctive. We find the three breeds each have a surprisingly different pattern of retrovirus insertions into their genomes compared with one another and with other UK breeds. Uniquely, there is a high incidence of the R0 retrotype in the Herdwick population, characteristic of a primitive genome found previously in very few breeds worldwide and none in the UK mainland. The Herdwick and Rough Fells carry two rare retroviral insertion events, common only in Texels, suggesting sheep populations in the northern uplands have a historical association with the original pin-tail sheep of Texel Island. Microsatellite data and analyses of SNPs associated with RXFP2 (horn traits) and PRLR (reproductive performance traits) also distinguished the three breeds. Significantly, an SNP linked to TMEM154, a locus controlling susceptibility to infection by Maedi-Visna, indicated that all three native hill breeds have a lower than average risk of infection to the lentivirus.
Conflict of interest statement
Figures
Similar articles
-
Recent advances in understanding the genetic resources of sheep breeds locally-adapted to the UK uplands: opportunities they offer for sustainable productivity.Front Genet. 2015 Feb 12;6:24. doi: 10.3389/fgene.2015.00024. eCollection 2015. Front Genet. 2015. PMID: 25729388 Free PMC article. Review.
-
First Report of SNPs Detection in TMEM154 Gene in Sheep from Poland and Their Association with SRLV Infection Status.Pathogens. 2024 Dec 30;14(1):16. doi: 10.3390/pathogens14010016. Pathogens. 2024. PMID: 39860977 Free PMC article.
-
The 1.78-kb insertion in the 3'-untranslated region of RXFP2 does not segregate with horn status in sheep breeds with variable horn status.Genet Sel Evol. 2016 Oct 19;48(1):78. doi: 10.1186/s12711-016-0256-3. Genet Sel Evol. 2016. PMID: 27760516 Free PMC article.
-
The genomic architecture of South African mutton, pelt, dual-purpose and nondescript sheep breeds relative to global sheep populations.Anim Genet. 2020 Dec;51(6):910-923. doi: 10.1111/age.12991. Epub 2020 Sep 7. Anim Genet. 2020. PMID: 32894610
-
Progress on ovine lentivirus and its resistant genes.Yi Chuan. 2014 Dec;36(12):1204-10. doi: 10.3724/SP.J.1005.2014.1204. Yi Chuan. 2014. PMID: 25487264 Review.
Cited by
-
Immunogenetics of small ruminant lentiviral infections.Viruses. 2014 Aug 22;6(8):3311-33. doi: 10.3390/v6083311. Viruses. 2014. PMID: 25153344 Free PMC article. Review.
-
Retroviral analysis reveals the ancient origin of Kihnu native sheep in Estonia: implications for breed conservation.Sci Rep. 2020 Oct 15;10(1):17340. doi: 10.1038/s41598-020-74415-z. Sci Rep. 2020. PMID: 33060653 Free PMC article.
-
Prospects in Innate Immune Responses as Potential Control Strategies against Non-Primate Lentiviruses.Viruses. 2018 Aug 17;10(8):435. doi: 10.3390/v10080435. Viruses. 2018. PMID: 30126090 Free PMC article. Review.
-
Paging through history: parchment as a reservoir of ancient DNA for next generation sequencing.Philos Trans R Soc Lond B Biol Sci. 2015 Jan 19;370(1660):20130379. doi: 10.1098/rstb.2013.0379. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 25487331 Free PMC article.
-
Recent advances in understanding the genetic resources of sheep breeds locally-adapted to the UK uplands: opportunities they offer for sustainable productivity.Front Genet. 2015 Feb 12;6:24. doi: 10.3389/fgene.2015.00024. eCollection 2015. Front Genet. 2015. PMID: 25729388 Free PMC article. Review.
References
-
- FAO (2007) The State of the world's animal genetic resources for food and agriculture. Pilling D, Rischkowsky B, editors. Rome.
-
- FAO (2009) Livestock keepers – guardians of biodiversity. Animal production and health paper no.167. Rome.
-
- Hoffmann I (2013) Adaptation to climate change – exploring the potential of locally adapted breeds. Animal 7: 346–362. - PubMed
-
- Toro M, Fernandez J, Caballero A (2009) Molecular characterization of breeds and its use in conservation. Livest Sci 120: 174–95.
-
- Groeneveld LF, Lenstra JA, Eding H, Toro MA, Scherf B, et al. (2010) Genetic diversity in farm animals – a review. Anim Genet 41: 6–31. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

