De novo design of self-assembling foldamers that inhibit heparin-protein interactions
- PMID: 24491145
- PMCID: PMC4324449
- DOI: 10.1021/cb500026x
De novo design of self-assembling foldamers that inhibit heparin-protein interactions
Abstract
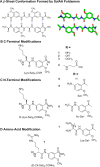
A series of self-associating foldamers have been designed as heparin reversal agents, as antidotes to prevent bleeding due to this potent antithrombotic agent. The foldamers have a repeating sequence of Lys-Sal, in which Sal is 5-amino-2-methoxy-benzoic acid. These foldamers are designed to self-associate along one face of an extended chain in a β-sheet-like interaction. The methoxy groups were included to form intramolecular hydrogen bonds that preclude the formation of very large amyloid-like aggregates, while the positively charged Lys side chains were introduced to interact electrostatically with the highly anionic heparin polymer. The prototype compound (Lys-Sal)4 carboxamide weakly associates in aqueous solution at physiological salt concentration in a monomer-dimer-hexamer equilibrium. The association is greatly enhanced at either high ionic strength or in the presence of a heparin derivative, which is bound tightly. Variants of this foldamer are active in an antithrombin III-factor Xa assay, showing their potential as heparin reversal agents.
Figures





References
-
- Gellman S. H. (1998) Foldamers: a manifesto. Acc. Chem. Res. 31, 173–180.
-
- Guichard G.; Huc I. (2011) Synthetic foldamers. Chem. Commun. 47, 5933–5941. - PubMed
-
- Hill D. J.; Mio M. J.; Prince R. B.; Hughes T. S.; Moore J. S. (2001) A field guide to foldamers. Chem. Rev. 101, 3893–4012. - PubMed
-
- Choi S.; Clements D. J.; Pophristic V.; Ivanov I.; Vemparala S.; Bennett J. S.; Klein M. L.; Winkler J. D.; DeGrado W. F. (2005) The design and evaluation of heparin-binding foldamers. Angew. Chem., Int. Ed. 44, 6685–6689. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

