Structural basis of efficient contagion: measles variations on a theme by parainfluenza viruses
- PMID: 24492202
- PMCID: PMC4028398
- DOI: 10.1016/j.coviro.2014.01.004
Structural basis of efficient contagion: measles variations on a theme by parainfluenza viruses
Abstract
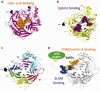
A quartet of attachment proteins and a trio of fusion protein subunits play the cell entry concert of parainfluenza viruses. While many of these viruses bind sialic acid to enter cells, wild type measles binds exclusively two tissue-specific proteins, the lymphatic receptor signaling lymphocytic activation molecule (SLAM), and the epithelial receptor nectin-4. SLAM binds near the stalk-head junction of the hemagglutinin. Nectin-4 binds a hydrophobic groove located between blades 4 and 5 of the hemagglutinin β-propeller head. The mutated vaccine strain hemagglutinin binds in addition the ubiquitous protein CD46, which explains attenuation. The measles virus entry concert has four movements. Andante misterioso: the virus takes over the immune system. Allegro con brio: it rapidly spreads in the upper airway's epithelia. 'Targeting' fugue: the versatile orchestra takes off. Presto furioso: the virus exits the host with thunder. Be careful: music is contagious.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures
References
-
- Griffin D. Measles. In: Knipe D, Howley P, editors. Fields’ Virology. Sixth Edition Vol. 1. Lippincott, Williams and Wilkins; 2013. pp. 1042–1069.
-
- Lamb RA, Parks G. Paramyxoviridae. In: Knipe D, Howley P, editors. Fields’ Virology. Sixth edition Vol. 1. Lippincott, Williams & Wilkins; 2013. pp. 957–995.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

