Consequences of glycan truncation on Fc structural integrity
- PMID: 24492344
- PMCID: PMC3896604
- DOI: 10.4161/mabs.26453
Consequences of glycan truncation on Fc structural integrity
Abstract
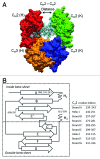
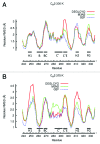
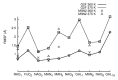
Effective characterization of protein-based therapeutic candidates such as monoclonal antibodies (mAbs) is important to facilitate their successful progression from early discovery and development stages to marketing approval. One challenge relevant to biopharmaceutical development is, understanding how the stability of a protein is affected by the presence of an attached oligosaccharide, termed a glycan. To explore the utility of molecular dynamics simulations as a complementary technique to currently available experimental methods, the Fc fragment was employed as a model system to improve our understanding of protein stabilization by glycan attachment. Long molecular dynamics simulations were performed on three Fc glycoform variants modeled using the crystal structure of a human IgG1 mAb. Two of these three glycoform variants have their glycan carbohydrates partially or completely removed. Structural differences among the glycoform variants during simulations suggest that glycan truncation and/or removal can cause quaternary structural deformation of the Fc as a result of the loss or disruption of a significant number of inter-glycan contacts that are not formed in the human IgG1 crystal structure, but do form during simulations described here. Glycan truncation/removal can also increase the tertiary structural deformation of CH2 domains, demonstrating the importance of specific carbohydrates toward stabilizing individual CH2 domains. At elevated temperatures, glycan truncation can also differentially affect structural deformation in locations (Helix-1 and Helix-2) that are far from the oligosaccharide attachment point. Deformation of these helices, which form part of the FcRn, could affect binding if these regions are unable to refold after temperature normalization. During elevated temperature simulations of the deglycosylated variant, CH2 domains collapsed onto CH3 domains. Observations from these glycan truncation/removal simulations have improved our understanding on how glycan composition can affect mAb stability.
Keywords: Fc; glycans; glycosylation; molecular dynamics; monoclonal antibodies; oligosaccharides; stability.
Figures



Similar articles
-
Effects of N-Glycan Composition on Structure and Dynamics of IgG1 Fc and Their Implications for Antibody Engineering.Sci Rep. 2017 Oct 4;7(1):12659. doi: 10.1038/s41598-017-12830-5. Sci Rep. 2017. PMID: 28978918 Free PMC article.
-
Crystal Structure of a Homogeneous IgG-Fc Glycoform with the N-Glycan Designed to Maximize the Antibody Dependent Cellular Cytotoxicity.ACS Chem Biol. 2017 May 19;12(5):1335-1345. doi: 10.1021/acschembio.7b00140. Epub 2017 Mar 31. ACS Chem Biol. 2017. PMID: 28318221
-
Molecular dynamics simulation of the crystallizable fragment of IgG1-insights for the design of Fcabs.Int J Mol Sci. 2014 Jan 2;15(1):438-55. doi: 10.3390/ijms15010438. Int J Mol Sci. 2014. PMID: 24451126 Free PMC article.
-
Glycosylation of IgG-Fc: a molecular perspective.Int Immunol. 2017 Jul 1;29(7):311-317. doi: 10.1093/intimm/dxx038. Int Immunol. 2017. PMID: 28655198 Review.
-
Antibody glycosylation and its impact on the pharmacokinetics and pharmacodynamics of monoclonal antibodies and Fc-fusion proteins.J Pharm Sci. 2015 Jun;104(6):1866-1884. doi: 10.1002/jps.24444. Epub 2015 Apr 14. J Pharm Sci. 2015. PMID: 25872915 Review.
Cited by
-
Rational design of viscosity reducing mutants of a monoclonal antibody: hydrophobic versus electrostatic inter-molecular interactions.MAbs. 2015;7(1):212-30. doi: 10.4161/19420862.2014.985504. MAbs. 2015. PMID: 25559441 Free PMC article.
-
The Impact of Immunoglobulin G1 Fc Sialylation on Backbone Amide H/D Exchange.Antibodies (Basel). 2019 Oct 1;8(4):49. doi: 10.3390/antib8040049. Antibodies (Basel). 2019. PMID: 31581521 Free PMC article.
-
Influence of N-glycosylation on effector functions and thermal stability of glycoengineered IgG1 monoclonal antibody with homogeneous glycoforms.MAbs. 2019 Feb/Mar;11(2):350-372. doi: 10.1080/19420862.2018.1551044. Epub 2018 Dec 10. MAbs. 2019. PMID: 30466347 Free PMC article.
-
Enhanced binding of antibodies generated during chronic HIV infection to mucus component MUC16.Mucosal Immunol. 2016 Nov;9(6):1549-1558. doi: 10.1038/mi.2016.8. Epub 2016 Mar 9. Mucosal Immunol. 2016. PMID: 26960182 Free PMC article.
-
Chemical Structure and Composition of Major Glycans Covalently Linked to Therapeutic Monoclonal Antibodies by Middle-Down Nuclear Magnetic Resonance.Anal Chem. 2018 Sep 18;90(18):11016-11024. doi: 10.1021/acs.analchem.8b02637. Epub 2018 Aug 27. Anal Chem. 2018. PMID: 30102512 Free PMC article.
References
-
- Kabat EA, Te Wu T, Gottesman KS, Foeller C. Sequences of proteins of immunological interest. Darby (PA); Diane Publishing; 1992.
-
- Raju S. Glycosylation variations with expression systems. Westborough (MA); BioProcess International; 2003:44-53.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
