Comparative analysis of proteomes and functionomes provides insights into origins of cellular diversification
- PMID: 24492748
- PMCID: PMC3892558
- DOI: 10.1155/2013/648746
Comparative analysis of proteomes and functionomes provides insights into origins of cellular diversification
Abstract
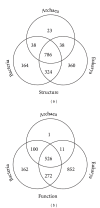
Reconstructing the evolutionary history of modern species is a difficult problem complicated by the conceptual and technical limitations of phylogenetic tree building methods. Here, we propose a comparative proteomic and functionomic inferential framework for genome evolution that allows resolving the tripartite division of cells and sketching their history. Evolutionary inferences were derived from the spread of conserved molecular features, such as molecular structures and functions, in the proteomes and functionomes of contemporary organisms. Patterns of use and reuse of these traits yielded significant insights into the origins of cellular diversification. Results uncovered an unprecedented strong evolutionary association between Bacteria and Eukarya while revealing marked evolutionary reductive tendencies in the archaeal genomic repertoires. The effects of nonvertical evolutionary processes (e.g., HGT, convergent evolution) were found to be limited while reductive evolution and molecular innovation appeared to be prevalent during the evolution of cells. Our study revealed a strong vertical trace in the history of proteins and associated molecular functions, which was reliably recovered using the comparative genomics approach. The trace supported the existence of a stem line of descent and the very early appearance of Archaea as a diversified superkingdom, but failed to uncover a hidden canonical pattern in which Bacteria was the first superkingdom to deploy superkingdom-specific structures and functions.
Figures


Similar articles
-
Reductive evolution of architectural repertoires in proteomes and the birth of the tripartite world.Genome Res. 2007 Nov;17(11):1572-85. doi: 10.1101/gr.6454307. Epub 2007 Oct 1. Genome Res. 2007. PMID: 17908824 Free PMC article.
-
The evolutionary history of protein fold families and proteomes confirms that the archaeal ancestor is more ancient than the ancestors of other superkingdoms.BMC Evol Biol. 2012 Jan 27;12:13. doi: 10.1186/1471-2148-12-13. BMC Evol Biol. 2012. PMID: 22284070 Free PMC article.
-
Global patterns of protein domain gain and loss in superkingdoms.PLoS Comput Biol. 2014 Jan 30;10(1):e1003452. doi: 10.1371/journal.pcbi.1003452. eCollection 2014 Jan. PLoS Comput Biol. 2014. PMID: 24499935 Free PMC article.
-
Archaea-First and the Co-Evolutionary Diversification of Domains of Life.Bioessays. 2018 Aug;40(8):e1800036. doi: 10.1002/bies.201800036. Epub 2018 Jun 26. Bioessays. 2018. PMID: 29944192 Review.
-
Transfer RNA and the origins of diversified life.Sci Prog. 2008;91(Pt 3):265-84. doi: 10.3184/003685008X360650. Sci Prog. 2008. PMID: 18853577 Free PMC article. Review.
Cited by
-
Horizontal gene transfer in human-associated microorganisms inferred by phylogenetic reconstruction and reconciliation.Sci Rep. 2019 Apr 11;9(1):5953. doi: 10.1038/s41598-019-42227-5. Sci Rep. 2019. PMID: 30976019 Free PMC article.
-
The natural history of molecular functions inferred from an extensive phylogenomic analysis of gene ontology data.PLoS One. 2017 May 3;12(5):e0176129. doi: 10.1371/journal.pone.0176129. eCollection 2017. PLoS One. 2017. PMID: 28467492 Free PMC article.
-
Arguments Reinforcing the Three-Domain View of Diversified Cellular Life.Archaea. 2016 Dec 5;2016:1851865. doi: 10.1155/2016/1851865. eCollection 2016. Archaea. 2016. PMID: 28050162 Free PMC article. Review.
-
Archaea: the first domain of diversified life.Archaea. 2014 Jun 2;2014:590214. doi: 10.1155/2014/590214. eCollection 2014. Archaea. 2014. PMID: 24987307 Free PMC article. Review.
-
Bacterial Origin and Reductive Evolution of the CPR Group.Genome Biol Evol. 2020 Mar 1;12(3):103-121. doi: 10.1093/gbe/evaa024. Genome Biol Evol. 2020. PMID: 32031619 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

