Genome recognition by MYC
- PMID: 24492846
- PMCID: PMC3904094
- DOI: 10.1101/cshperspect.a014191
Genome recognition by MYC
Abstract
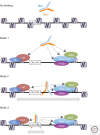
MYC dimerizes with MAX to bind DNA, with a preference for the E-box consensus CACGTG and several variant motifs. In cells, MYC binds DNA preferentially within transcriptionally active promoter regions. Although several thousand promoters are bound under physiological (low MYC) conditions, these represent only a fraction of all accessible, active promoters. MYC overexpression-as commonly observed in cancer cells-leads to invasion of virtually all active promoters, as well as of distal enhancer elements. We summarize here what is currently known about the mechanisms that may guide this process. We propose that binding site recognition is determined by low-affinity protein-protein interactions between MYC/MAX dimers and components of the basal transcriptional machinery, other chromatin-associated protein complexes, and/or DNA-bound transcription factors. DNA binding occurs subsequently, without an obligate requirement for sequence recognition. Local DNA scanning then leads to preferential stabilization of the MYC/MAX dimer on high-affinity DNA elements. This model is consistent with the invasion of all active promoters that occurs at elevated MYC levels, but posits that important differences in affinity persist between physiological target sites and the newly invaded elements, which may not all be bound in a productive regulatory mode. The implications of this model for transcriptional control by MYC in normal and cancer cells are discussed in the light of the latest literature.
Figures
References
-
- Adhikary S, Eilers M 2005. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 6: 635–645 - PubMed
-
- Amati B, Dalton S, Brooks MW, Littlewood TD, Evan GI, Land H 1992. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature 359: 423–426 - PubMed
-
- Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI, Land H 1993a. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell 72: 233–245 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
