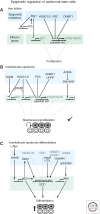
Epigenetic regulation of epidermal differentiation
- PMID: 24492849
- PMCID: PMC3904097
- DOI: 10.1101/cshperspect.a015263
Epigenetic regulation of epidermal differentiation
Abstract
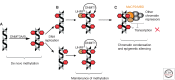
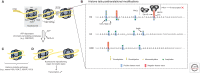
In a cell, the chromatin state is controlled by the highly regulated interplay of epigenetic mechanisms ranging from DNA methylation and incorporation of different histone variants to posttranslational modification of histones and ATP-dependent chromatin remodeling. These changes alter the structure of the chromatin to either facilitate or restrict the access of transcription machinery to DNA. These epigenetic modifications function to exquisitely orchestrate the expression of different genes, and together constitute the epigenome of a cell. In the skin, different epigenetic regulators form a regulatory network that operates to guarantee skin stem cell maintenance while controlling differentiation to multiple skin structures. In this review, we will discuss recent findings on epigenetic mechanisms of skin control and their relationship to skin pathologies.
Figures
References
-
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734 - PubMed
-
- Allis C, Berger S, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. 2007. New nomenclature for chromatin-modifying enzymes. Cell 131: 633–636 - PubMed
-
- Andrews AJ, Luger K 2011. Nucleosome structure(s) and stability: Variations on a theme. Annu Rev Biophys 40: 99–117 - PubMed
-
- Bao Y 2011. Chromatin response to DNA double-strand break damage. Epigenomics 3: 307–321 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
