SegAnnDB: interactive Web-based genomic segmentation
- PMID: 24493034
- PMCID: PMC4029035
- DOI: 10.1093/bioinformatics/btu072
SegAnnDB: interactive Web-based genomic segmentation
Abstract
Motivation: DNA copy number profiles characterize regions of chromosome gains, losses and breakpoints in tumor genomes. Although many models have been proposed to detect these alterations, it is not clear which model is appropriate before visual inspection the signal, noise and models for a particular profile.
Results: We propose SegAnnDB, a Web-based computer vision system for genomic segmentation: first, visually inspect the profiles and manually annotate altered regions, then SegAnnDB determines the precise alteration locations using a mathematical model of the data and annotations. SegAnnDB facilitates collaboration between biologists and bioinformaticians, and uses the University of California, Santa Cruz genome browser to visualize copy number alterations alongside known genes.
Availability and implementation: The breakpoints project on INRIA GForge hosts the source code, an Amazon Machine Image can be launched and a demonstration Web site is http://bioviz.rocq.inria.fr.
© The Author 2014. Published by Oxford University Press.
Figures

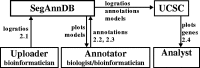

 black scale, the breakpoints in the segmentation model are red and the inferred copy number status of each segment is shown using the copy number annotation colors of
black scale, the breakpoints in the segmentation model are red and the inferred copy number status of each segment is shown using the copy number annotation colors of 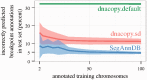

References
-
- Bostock M, et al. D3 data-driven documents. IEEE Trans. Vis. Comput. Graph. 2011;17:2301–2309. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

