Long noncoding RNA in liver diseases
- PMID: 24493213
- PMCID: PMC4110118
- DOI: 10.1002/hep.27043
Long noncoding RNA in liver diseases
Abstract
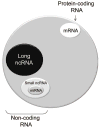
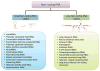
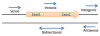
The identification of the presence of large RNA transcripts that do not code for proteins but that may have biological functions has provided an important new perspective in gene regulation. These long noncoding RNAs (lncRNAs) are being increasingly recognized to contribute to many biological processes through diverse mechanisms. The roles of these emerging genes are being recognized across kingdoms. These findings are profoundly altering our understanding of disease pathobiology and leading to the emergence of new biological concepts underlying human diseases. Strategies for the discovery and characterization of lncRNAs are highlighted. Several lncRNAs have been described in liver disease, and in liver cancers in particular. Their molecular mechanisms of action, function, and contributions to disease pathophysiology are reviewed. LncRNA genes associated with liver diseases have potential roles as biomarkers of disease diagnosis, prognosis, or therapeutic response as well as direct targets for therapeutic intervention.
Conclusion: The emerging knowledge in this rapidly advancing field offers promise for new fundamental knowledge and clinical applications that will be relevant for human liver diseases.
© 2014 by the American Association for the Study of Liver Diseases.
Figures


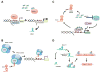
References
-
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
