Gene expression signature for biliary atresia and a role for interleukin-8 in pathogenesis of experimental disease
- PMID: 24493287
- PMCID: PMC4077977
- DOI: 10.1002/hep.27045
Gene expression signature for biliary atresia and a role for interleukin-8 in pathogenesis of experimental disease
Abstract
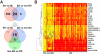
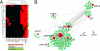
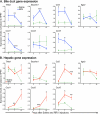
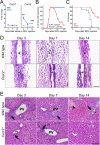
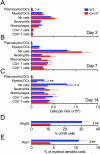
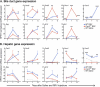
Biliary atresia (BA) is a progressive fibroinflammatory obstruction of extrahepatic bile ducts that presents as neonatal cholestasis. Due to the overlap in clinical, biochemical, and histological features with other causes of cholestasis, the diagnosis requires an intraoperative cholangiogram. Thus, we determined whether diseased livers express a gene expression signature unique to BA. Applying stringent statistical analysis to a genome-wide liver expression platform of 64 infants with BA at the time of diagnosis, 14 age-appropriate subjects with intrahepatic cholestasis as diseased controls and seven normal controls, we identified 15 genes uniquely expressed in BA with an accuracy of 92.3%. Among these genes, IL8 and LAMC2 were sufficient to classify subjects with BA distinctly from diseased controls with an area under the curve of 0.934 (95% confidence interval [CI]: 0.84-1.03), sensitivity of 96.9%, and specificity of 85.7% using their combined first principal component. Direct measurement of interleukin (IL)8 protein in the serum, however, was not different between the two groups. To investigate whether the liver-restricted increase in IL8 was relevant to disease pathogenesis, we inactivated the signaling of IL8 homologs by genetic targeting of the Cxcr2 receptor in a murine model of experimental BA. Disruption of Cxcr2 shortened the duration of cholestasis, decreased the incidence of bile duct obstruction, and improved survival above wild-type neonatal mice.
Conclusion: The hepatic expression of IL8 and LAMC2 has high sensitivity for BA at diagnosis and may serve as a biomarker of disease, with an important role for the IL8 signaling in experimental BA.
Trial registration: ClinicalTrials.gov NCT00061828.
© 2014 by the American Association for the Study of Liver Diseases.
Figures

Comment in
-
Hepatic expression of interleukin 8 in diagnosing biliary atresia.Hepatology. 2015 Feb;61(2):731-2. doi: 10.1002/hep.27229. Epub 2014 Jul 31. Hepatology. 2015. PMID: 24841829 No abstract available.
-
Reply: To PMID 24493287.Hepatology. 2015 Feb;61(2):732-3. doi: 10.1002/hep.27231. Epub 2014 Jul 31. Hepatology. 2015. PMID: 24844795 No abstract available.
References
-
- Moyer V, Freese DK, Whitington PF, Olson AD, Brewer F, Colletti RB, Heyman MB. Guideline for the evaluation of cholestatic jaundice in infants: recommendations of the North American Society for Pediatric Gastroenterology, Hepatology and Nutrition. J Pediatr Gastroenterol Nutr. 2004;39:115–128. - PubMed
-
- Bezerra JA, Tiao G, Ryckman FC, Alonso M, Sabla GE, Shneider B, Sokol RJ, et al. Genetic induction of proinflammatory immunity in children with biliary atresia. Lancet. 2002;360:1653–1659. - PubMed
-
- Ohya T, Fujimoto T, Shimomura H, Miyano T. Degeneration of intrahepatic bile duct with lymphocyte infiltration into biliary epithelial cells in biliary atresia. J Pediatr Surg. 1995;30:515–518. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- R01 DK83781/DK/NIDDK NIH HHS/United States
- 8 UL1 TR000077-04/TR/NCATS NIH HHS/United States
- DK62497/DK/NIDDK NIH HHS/United States
- U01 DK084536/DK/NIDDK NIH HHS/United States
- U01 DK062456/DK/NIDDK NIH HHS/United States
- R01 DK083781/DK/NIDDK NIH HHS/United States
- DK78392/DK/NIDDK NIH HHS/United States
- U01 DK062497/DK/NIDDK NIH HHS/United States
- P30 DK078392/DK/NIDDK NIH HHS/United States
- U01 DK103135/DK/NIDDK NIH HHS/United States
- R01 DK064008/DK/NIDDK NIH HHS/United States
- UL1 TR000077/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
