Design and application of cotranscriptional non-enzymatic RNA circuits and signal transducers
- PMID: 24493736
- PMCID: PMC3985647
- DOI: 10.1093/nar/gku074
Design and application of cotranscriptional non-enzymatic RNA circuits and signal transducers
Abstract
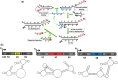
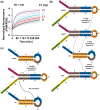
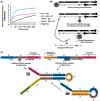
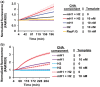
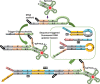
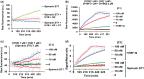
Nucleic acid circuits are finding increasing real-life applications in diagnostics and synthetic biology. Although DNA has been the main operator in most nucleic acid circuits, transcriptionally produced RNA circuits could provide powerful alternatives for reagent production and their use in cells. Towards these goals, we have implemented a particular nucleic acid circuit, catalytic hairpin assembly, using RNA for both information storage and processing. Our results demonstrated that the design principles developed for DNA circuits could be readily translated to engineering RNA circuits that operated with similar kinetics and sensitivities of detection. Not only could purified RNA hairpins perform amplification reactions but RNA hairpins transcribed in vitro also mediated amplification, even without purification. Moreover, we could read the results of the non-enzymatic amplification reactions using a fluorescent RNA aptamer 'Spinach' that was engineered to undergo sequence-specific conformational changes. These advances were applied to the end-point and real-time detection of the isothermal strand displacement amplification reaction that produces single-stranded DNAs as part of its amplification cycle. We were also able to readily engineer gate structures with RNA similar to those that have previously formed the basis of DNA circuit computations. Taken together, these results validate an entirely new chemistry for the implementation of nucleic acid circuits.
Figures



References
-
- Qian L, Winfree E, Bruck J. Neural network computation with DNA strand displacement cascades. Nature. 2011;475:368–372. - PubMed
-
- Zhang DY, Seelig G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 2011;3:103–113. - PubMed
-
- Qian L, Winfree E. Scaling up digital circuit computation with DNA strand displacement cascades. Science. 2011;332:1196–1201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

