Investigating the mechanisms of ribonucleotide excision repair in Escherichia coli
- PMID: 24495324
- PMCID: PMC4089967
- DOI: 10.1016/j.mrfmmm.2014.01.005
Investigating the mechanisms of ribonucleotide excision repair in Escherichia coli
Abstract
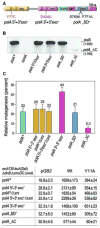
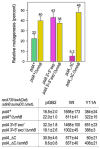
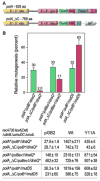
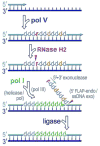
Low fidelity Escherichia coli DNA polymerase V (pol V/UmuD'2C) is best characterized for its ability to perform translesion synthesis (TLS). However, in recA730 lexA(Def) strains, the enzyme is expressed under optimal conditions allowing it to compete with the cell's replicase for access to undamaged chromosomal DNA and leads to a substantial increase in spontaneous mutagenesis. We have recently shown that a Y11A substitution in the "steric gate" residue of UmuC reduces both base and sugar selectivity of pol V, but instead of generating an increased number of spontaneous mutations, strains expressing umuC_Y11A are poorly mutable in vivo. This phenotype is attributed to efficient RNase HII-initiated repair of the misincorporated ribonucleotides that concomitantly removes adjacent misincorporated deoxyribonucleotides. We have utilized the ability of the pol V steric gate mutant to promote incorporation of large numbers of errant ribonucleotides into the E. coli genome to investigate the fundamental mechanisms underlying ribonucleotide excision repair (RER). Here, we demonstrate that RER is normally facilitated by DNA polymerase I (pol I) via classical "nick translation". In vitro, pol I displaces 1-3 nucleotides of the RNA/DNA hybrid and through its 5'→3' (exo/endo) nuclease activity releases ribo- and deoxyribonucleotides from DNA. In vivo, umuC_Y11A-dependent mutagenesis changes significantly in polymerase-deficient, or proofreading-deficient polA strains, indicating a pivotal role for pol I in ribonucleotide excision repair (RER). However, there is also considerable redundancy in the RER pathway in E. coli. Pol I's strand displacement and FLAP-exo/endonuclease activities can be facilitated by alternate enzymes, while the DNA polymerization step can be assumed by high-fidelity pol III. We conclude that RNase HII and pol I normally act to minimize the genomic instability that is generated through errant ribonucleotide incorporation, but that the "nick-translation" activities encoded by the single pol I polypeptide can be undertaken by a variety of back-up enzymes.
Keywords: RNase H; Ribonucleotide excision repair; Steric gate mutant; UmuC: spontaneous mutagenesis; Y-family DNA polymerase.
Published by Elsevier B.V.
Conflict of interest statement
The authors declare that there is no conflict of interest
Figures


Similar articles
-
Mechanisms employed by Escherichia coli to prevent ribonucleotide incorporation into genomic DNA by Pol V.PLoS Genet. 2012;8(11):e1003030. doi: 10.1371/journal.pgen.1003030. Epub 2012 Nov 8. PLoS Genet. 2012. PMID: 23144626 Free PMC article.
-
Role of RNase H enzymes in maintaining genome stability in Escherichia coli expressing a steric-gate mutant of pol VICE391.DNA Repair (Amst). 2019 Dec;84:102685. doi: 10.1016/j.dnarep.2019.102685. Epub 2019 Aug 10. DNA Repair (Amst). 2019. PMID: 31543434 Free PMC article.
-
Removal of misincorporated ribonucleotides from prokaryotic genomes: an unexpected role for nucleotide excision repair.PLoS Genet. 2013 Nov;9(11):e1003878. doi: 10.1371/journal.pgen.1003878. Epub 2013 Nov 7. PLoS Genet. 2013. PMID: 24244177 Free PMC article.
-
Redundancy in ribonucleotide excision repair: Competition, compensation, and cooperation.DNA Repair (Amst). 2015 May;29:74-82. doi: 10.1016/j.dnarep.2015.02.008. Epub 2015 Feb 16. DNA Repair (Amst). 2015. PMID: 25753809 Free PMC article. Review.
-
Mutations for Worse or Better: Low-Fidelity DNA Synthesis by SOS DNA Polymerase V Is a Tightly Regulated Double-Edged Sword.Biochemistry. 2016 Apr 26;55(16):2309-18. doi: 10.1021/acs.biochem.6b00117. Epub 2016 Apr 12. Biochemistry. 2016. PMID: 27043933 Free PMC article. Review.
Cited by
-
RNase HIII Is Important for Okazaki Fragment Processing in Bacillus subtilis.J Bacteriol. 2019 Mar 13;201(7):e00686-18. doi: 10.1128/JB.00686-18. Print 2019 Apr 1. J Bacteriol. 2019. PMID: 30670546 Free PMC article.
-
How the misincorporation of ribonucleotides into genomic DNA can be both harmful and helpful to cells.Nucleic Acids Res. 2014;42(16):10226-34. doi: 10.1093/nar/gku773. Epub 2014 Aug 26. Nucleic Acids Res. 2014. PMID: 25159610 Free PMC article. Review.
-
Mutagenic cost of ribonucleotides in bacterial DNA.Proc Natl Acad Sci U S A. 2017 Oct 31;114(44):11733-11738. doi: 10.1073/pnas.1710995114. Epub 2017 Oct 16. Proc Natl Acad Sci U S A. 2017. PMID: 29078353 Free PMC article.
-
Ribonucleotide incorporation into DNA during DNA replication and its consequences.Crit Rev Biochem Mol Biol. 2021 Feb;56(1):109-124. doi: 10.1080/10409238.2020.1869175. Epub 2021 Jan 18. Crit Rev Biochem Mol Biol. 2021. PMID: 33461360 Free PMC article. Review.
-
Determinants of spontaneous mutation in the bacterium Escherichia coli as revealed by whole-genome sequencing.Proc Natl Acad Sci U S A. 2015 Nov 3;112(44):E5990-9. doi: 10.1073/pnas.1512136112. Epub 2015 Oct 12. Proc Natl Acad Sci U S A. 2015. PMID: 26460006 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

