Comparison of three next-generation sequencing platforms for metagenomic sequencing and identification of pathogens in blood
- PMID: 24495417
- PMCID: PMC3922542
- DOI: 10.1186/1471-2164-15-96
Comparison of three next-generation sequencing platforms for metagenomic sequencing and identification of pathogens in blood
Abstract
Background: The introduction of benchtop sequencers has made adoption of whole genome sequencing possible for a broader community of researchers than ever before. Concurrently, metagenomic sequencing (MGS) is rapidly emerging as a tool for interrogating complex samples that defy conventional analyses. In addition, next-generation sequencers are increasingly being used in clinical or related settings, for instance to track outbreaks. However, information regarding the analytical sensitivity or limit of detection (LoD) of benchtop sequencers is currently lacking. Furthermore, the specificity of sequence information at or near the LoD is unknown.
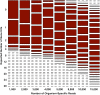
Results: In the present study, we assess the ability of three next-generation sequencing platforms to identify a pathogen (viral or bacterial) present in low titers in a clinically relevant sample (blood). Our results indicate that the Roche-454 Titanium platform is capable of detecting Dengue virus at titers as low as 1X102.5 pfu/mL, corresponding to an estimated 5.4X104 genome copies/ml maximum. The increased throughput of the benchtop sequencers, the Ion Torrent PGM and Illumina MiSeq platforms, enabled detection of viral genomes at concentrations as low as 1X104 genome copies/mL. Platform-specific biases were evident in sequence read distributions as well as viral genome coverage. For bacterial samples, only the MiSeq platform was able to provide sequencing reads that could be unambiguously classified as originating from Bacillus anthracis.
Conclusion: The analytical sensitivity of all three platforms approaches that of standard qPCR assays. Although all platforms were able to detect pathogens at the levels tested, there were several noteworthy differences. The Roche-454 Titanium platform produced consistently longer reads, even when compared with the latest chemistry updates for the PGM platform. The MiSeq platform produced consistently greater depth and breadth of coverage, while the Ion Torrent was unequaled for speed of sequencing. None of the platforms were able to verify a single nucleotide polymorphism responsible for antiviral resistance in an Influenza A strain isolated from the 2009 H1N1 pandemic. Overall, the benchtop platforms perform well for identification of pathogens from a representative clinical sample. However, unlike identification, characterization of pathogens is likely to require higher titers, multiple libraries and/or multiple sequencing runs.
Figures


Similar articles
-
Analysis of the genetic diversity of influenza A viruses using next-generation DNA sequencing.BMC Genomics. 2015 Feb 14;16(1):79. doi: 10.1186/s12864-015-1284-z. BMC Genomics. 2015. PMID: 25758772 Free PMC article.
-
Comparison of Illumina MiSeq and the Ion Torrent PGM and S5 platforms for whole-genome sequencing of picornaviruses and caliciviruses.J Virol Methods. 2020 Jun;280:113865. doi: 10.1016/j.jviromet.2020.113865. Epub 2020 Apr 14. J Virol Methods. 2020. PMID: 32302601 Free PMC article.
-
A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers.BMC Genomics. 2012 Jul 24;13:341. doi: 10.1186/1471-2164-13-341. BMC Genomics. 2012. PMID: 22827831 Free PMC article.
-
MiSeq: A Next Generation Sequencing Platform for Genomic Analysis.Methods Mol Biol. 2018;1706:223-232. doi: 10.1007/978-1-4939-7471-9_12. Methods Mol Biol. 2018. PMID: 29423801 Review.
-
HLA typing by next-generation sequencing - getting closer to reality.Tissue Antigens. 2014 Feb;83(2):65-75. doi: 10.1111/tan.12298. Tissue Antigens. 2014. PMID: 24447174 Review.
Cited by
-
Novel Virus Identification through Metagenomics: A Systematic Review.Life (Basel). 2022 Dec 7;12(12):2048. doi: 10.3390/life12122048. Life (Basel). 2022. PMID: 36556413 Free PMC article. Review.
-
Bacillus-Dominant Airborne Bacterial Communities Identified During Asian Dust Events.Microb Ecol. 2019 Oct;78(3):677-687. doi: 10.1007/s00248-019-01348-0. Epub 2019 Mar 23. Microb Ecol. 2019. PMID: 30904989
-
Perspective for Aquaponic Systems: "Omic" Technologies for Microbial Community Analysis.Biomed Res Int. 2015;2015:480386. doi: 10.1155/2015/480386. Epub 2015 Oct 5. Biomed Res Int. 2015. PMID: 26509157 Free PMC article. Review.
-
Efficient and unbiased metagenomic recovery of RNA virus genomes from human plasma samples.Sci Rep. 2017 Jun 23;7(1):4173. doi: 10.1038/s41598-017-02239-5. Sci Rep. 2017. PMID: 28646219 Free PMC article.
-
A tale of two approaches: how metagenomics and proteomics are shaping the future of encephalitis diagnostics.Curr Opin Neurol. 2015 Jun;28(3):283-7. doi: 10.1097/WCO.0000000000000198. Curr Opin Neurol. 2015. PMID: 25923127 Free PMC article. Review.
References
-
- Moore RA, Warren RL, Freeman JD, Gustavsen JA, Chenard C, Friedman JM, Suttle CA, Zhao Y, Holt RA. The sensitivity of massively parallel sequencing for detecting candidate infectious agents associated with human tissue. PLoS One. 2011;15(5):e19838. doi: 10.1371/journal.pone.0019838. - DOI - PMC - PubMed
-
- Cheval J, Sauvage V, Frangeul L, Dacheux L, Guigon G, Dumey N, Pariente K, Rousseaux C, Dorange F, Berthet N. et al.Evaluation of high-throughput sequencing for identifying known and unknown viruses in biological samples. J Clin Microbiol. 2011;15(9):3268–3275. doi: 10.1128/JCM.00850-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

