Zip nucleic acid: a new reliable method to increase the melting temperature of real-time PCR probes
- PMID: 24495816
- PMCID: PMC3922743
- DOI: 10.1186/2251-6581-13-26
Zip nucleic acid: a new reliable method to increase the melting temperature of real-time PCR probes
Abstract
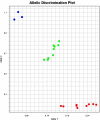
TaqMan genotyping with real-time PCR is a reliable method for single nucleotide polymorphism detection, which is done by probes. These oligonucleotides should be short enough to avoid mismatch hybridization, as well as having 5-10°C higher melting temperature than the primers of real-time PCR reaction. One approach for these qualities is to conjugate the probe with minor groove binder (MGB). Having no access to MGB probes, we searched for an alternative. In the current study, we used Zip Nucleic Acids (ZNA) as probes to increase its stability and melting temperature. Our aim was to genotype the -265 T/C changes of Apolipoprotein A-2 gene. We set up the real-time PCR reaction with ZNA probes, and by repeating the reactions, we confirmed the reliability of this new approach. It is now recommended to use ZNA probes, as an alternative to MGB probes, to increase the probe Tm value and its binding to target DNA.
Figures

Similar articles
-
[Research on ZNA probe in the quantitative detection of chlamydia trachomatis nucleic acid].Zhonghua Yu Fang Yi Xue Za Zhi. 2020 Oct 6;54(10):1133-1140. doi: 10.3760/cma.j.cn112150-20200626-00928. Zhonghua Yu Fang Yi Xue Za Zhi. 2020. PMID: 33115201 Chinese.
-
Single nucleotide polymorphism genotyping by two colour melting curve analysis using the MGB Eclipse Probe System in challenging sequence environment.Hum Genomics. 2004 Mar;1(3):209-17. doi: 10.1186/1479-7364-1-3-209. Hum Genomics. 2004. PMID: 15588480 Free PMC article.
-
Zip nucleic acids are potent hydrolysis probes for quantitative PCR.Nucleic Acids Res. 2010 Apr;38(7):e95. doi: 10.1093/nar/gkp1218. Epub 2010 Jan 13. Nucleic Acids Res. 2010. PMID: 20071749 Free PMC article.
-
Real-time PCR detection chemistry.Clin Chim Acta. 2015 Jan 15;439:231-50. doi: 10.1016/j.cca.2014.10.017. Epub 2014 Oct 22. Clin Chim Acta. 2015. PMID: 25451956 Review.
-
Single nucleotide polymorphism genotyping using locked nucleic acid (LNA).Expert Rev Mol Diagn. 2003 Jan;3(1):27-38. doi: 10.1586/14737159.3.1.27. Expert Rev Mol Diagn. 2003. PMID: 12528362 Review.
Cited by
-
The interaction between ApoA2 -265T>C polymorphism and dietary fatty acids intake on oxidative stress in patients with type 2 diabetes mellitus.Eur J Nutr. 2017 Aug;56(5):1931-1938. doi: 10.1007/s00394-016-1235-8. Epub 2016 Jun 6. Eur J Nutr. 2017. PMID: 27271094
-
Interaction between Apo A-II -265T>C polymorphism and dietary total antioxidant capacity on some anthropometric indices and serum lipid profile in patients with type 2 diabetes mellitus.J Nutr Sci. 2021 Feb 9;10:e9. doi: 10.1017/jns.2020.61. eCollection 2021. J Nutr Sci. 2021. PMID: 33889392 Free PMC article.
-
Electrochemical Detection of Solution Phase Hybridization Related to Single Nucleotide Mutation by Carbon Nanofibers Enriched Electrodes.Materials (Basel). 2019 Oct 16;12(20):3377. doi: 10.3390/ma12203377. Materials (Basel). 2019. PMID: 31623126 Free PMC article.
-
Dietary acid load modifies the effects of ApoA2-265 T > C polymorphism on lipid profile and serum leptin and ghrelin levels among type 2 diabetic patients.BMC Endocr Disord. 2022 Jul 26;22(1):190. doi: 10.1186/s12902-022-01083-7. BMC Endocr Disord. 2022. PMID: 35883173 Free PMC article.
-
Impedimetric Sensing of Factor V Leiden Mutation by Zip Nucleic Acid Probe and Electrochemical Array.Biosensors (Basel). 2020 Sep 7;10(9):116. doi: 10.3390/bios10090116. Biosensors (Basel). 2020. PMID: 32906640 Free PMC article.
References
-
- Pestana E, Belak S, Diallo A, Crowther J, Viljoen G. Early, Rapid and Sensitive Veterinary Molecular Diagnostics - Real Time PCR Applications. London: Springer; 2010.
-
- Kutyavin IV, Afonina IA, Mills A, Gorn VV, Lukhtanov EA, Belousov ES, Singer MJ, Walburger DK, Lokhov SG, Gall AA, Dempcy R, Reed MW, Meyer RB, Hedgpeth J. 3′-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000;28:655–661. doi: 10.1093/nar/28.2.655. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

