In vitro Inactivation of Latent HSV by Targeted Mutagenesis Using an HSV-specific Homing Endonuclease
- PMID: 24496438
- PMCID: PMC3951911
- DOI: 10.1038/mtna.2013.75
In vitro Inactivation of Latent HSV by Targeted Mutagenesis Using an HSV-specific Homing Endonuclease
Abstract
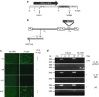
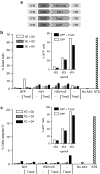
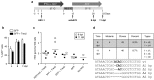
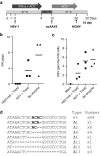
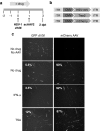
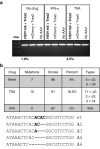
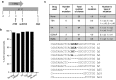
Following acute infection, herpes simplex virus (HSV) establishes latency in sensory neurons, from which it can reactivate and cause recurrent disease. Available antiviral therapies do not affect latent viral genomes; therefore, they do not prevent reactivation following therapy cessation. One possible curative approach involves the introduction of DNA double strand breaks in latent HSV genomes by rare-cutting endonucleases, leading to mutagenesis of essential viral genes. We tested this approach in an in vitro HSV latency model using the engineered homing endonuclease (HE) HSV1m5, which recognizes a sequence in the HSV-1 gene UL19, encoding the virion protein VP5. Coexpression of the 3'-exonuclease Trex2 with HEs increased HE-mediated mutagenesis frequencies up to sixfold. Following HSV1m5/Trex2 delivery with adeno-associated viral (AAV) vectors, the target site was mutated in latent HSV genomes with no detectable cell toxicity. Importantly, HSV production by latently infected cells after reactivation was decreased after HSV1m5/Trex2 exposure. Exposure to histone deacetylase inhibitors prior to HSV1m5/Trex2 treatment increased mutagenesis frequencies of latent HSV genomes another two- to fivefold, suggesting that chromatin modification may be a useful adjunct to gene-targeting approaches. These results support the continuing development of HEs and other nucleases (ZFNs, TALENs, CRISPRs) for cure of chronic viral infections.Molecular Therapy-Nucleic Acids (2014) 3, e1; doi:10.1038/mtna.2013.75; published online 4 February 2014.
Figures
References
-
- Xu F, Sternberg MR, Kottiri BJ, McQuillan GM, Lee FK, Nahmias AJ, et al. Trends in herpes simplex virus type 1 and type 2 seroprevalence in the United States. JAMA. 2006;296:964–973. - PubMed
-
- Brown EL, Gardella C, Malm G, Prober CG, Forsgren M, Krantz EM, et al. Effect of maternal herpes simplex virus (HSV) serostatus and HSV type on risk of neonatal herpes. Acta Obstet Gynecol Scand. 2007;86:523–529. - PubMed
-
- Serwadda D, Gray RH, Sewankambo NK, Wabwire-Mangen F, Chen MZ, Quinn TC, et al. Human immunodeficiency virus acquisition associated with genital ulcer disease and herpes simplex virus type 2 infection: a nested case-control study in Rakai, Uganda. J Infect Dis. 2003;188:1492–1497. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

