Differential methylation of the TRPA1 promoter in pain sensitivity
- PMID: 24496475
- PMCID: PMC3926001
- DOI: 10.1038/ncomms3978
Differential methylation of the TRPA1 promoter in pain sensitivity
Abstract
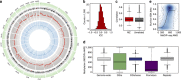
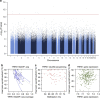
Chronic pain is a global public health problem, but the underlying molecular mechanisms are not fully understood. Here we examine genome-wide DNA methylation, first in 50 identical twins discordant for heat pain sensitivity and then in 50 further unrelated individuals. Whole-blood DNA methylation was characterized at 5.2 million loci by MeDIP sequencing and assessed longitudinally to identify differentially methylated regions associated with high or low pain sensitivity (pain DMRs). Nine meta-analysis pain DMRs show robust evidence for association (false discovery rate 5%) with the strongest signal in the pain gene TRPA1 (P=1.2 × 10(-13)). Several pain DMRs show longitudinal stability consistent with susceptibility effects, have similar methylation levels in the brain and altered expression in the skin. Our approach identifies epigenetic changes in both novel and established candidate genes that provide molecular insights into pain and may generalize to other complex traits.
Conflict of interest statement
Authors A.K.L., B.Z., C.L.H., S.S. and S.L.J. are employees of Pfizer Inc. All other authors declare no competing financial interest.
Figures

References
-
- Breivik H., Collett B., Ventafridda V., Cohen R. & Gallacher D. Survey of chronic pain in Europe: prevalence, impact on daily life, and treatment. Eur. J. Pain 10, 287–333 (2006). - PubMed
-
- Nielsen C. S., Price D. D., Vassend O., Stubhaug A. & Harris J. R. Characterizing individual differences in heat-pain sensitivity. Pain 119, 65–74 (2005). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

