Machine learning classification of cell-specific cardiac enhancers uncovers developmental subnetworks regulating progenitor cell division and cell fate specification
- PMID: 24496624
- PMCID: PMC3912831
- DOI: 10.1242/dev.101709
Machine learning classification of cell-specific cardiac enhancers uncovers developmental subnetworks regulating progenitor cell division and cell fate specification
Abstract
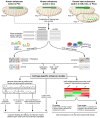
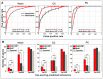
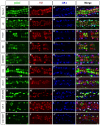
The Drosophila heart is composed of two distinct cell types, the contractile cardial cells (CCs) and the surrounding non-muscle pericardial cells (PCs), development of which is regulated by a network of conserved signaling molecules and transcription factors (TFs). Here, we used machine learning with array-based chromatin immunoprecipitation (ChIP) data and TF sequence motifs to computationally classify cell type-specific cardiac enhancers. Extensive testing of predicted enhancers at single-cell resolution revealed the added value of ChIP data for modeling cell type-specific activities. Furthermore, clustering the top-scoring classifier sequence features identified novel cardiac and cell type-specific regulatory motifs. For example, we found that the Myb motif learned by the classifier is crucial for CC activity, and the Myb TF acts in concert with two forkhead domain TFs and Polo kinase to regulate cardiac progenitor cell divisions. In addition, differential motif enrichment and cis-trans genetic studies revealed that the Notch signaling pathway TF Suppressor of Hairless [Su(H)] discriminates PC from CC enhancer activities. Collectively, these studies elucidate molecular pathways used in the regulatory decisions for proliferation and differentiation of cardiac progenitor cells, implicate Su(H) in regulating cell fate decisions of these progenitors, and document the utility of enhancer modeling in uncovering developmental regulatory subnetworks.
Keywords: Cell division; Drosophila; Gene regulation; Machine learning; Organogenesis; Progenitor specification; Transcription factors.
Figures





Similar articles
-
Three distinct mechanisms, Notch instructive, permissive, and independent, regulate the expression of two different pericardial genes to specify cardiac cell subtypes.PLoS One. 2020 Oct 27;15(10):e0241191. doi: 10.1371/journal.pone.0241191. eCollection 2020. PLoS One. 2020. PMID: 33108408 Free PMC article.
-
Myogenic cells fates are antagonized by Notch only in asymmetric lineages of the Drosophila heart, with or without cell division.Development. 2003 Jul;130(13):3039-51. doi: 10.1242/dev.00484. Development. 2003. PMID: 12756185
-
Enhancer modeling uncovers transcriptional signatures of individual cardiac cell states in Drosophila.Nucleic Acids Res. 2015 Feb 18;43(3):1726-39. doi: 10.1093/nar/gkv011. Epub 2015 Jan 21. Nucleic Acids Res. 2015. PMID: 25609699 Free PMC article.
-
Enhancers: emerging roles in cell fate specification.EMBO Rep. 2012 Apr 10;13(5):423-30. doi: 10.1038/embor.2012.52. EMBO Rep. 2012. PMID: 22491032 Free PMC article. Review.
-
Drosophila myogenesis and insights into the role of nautilus.Curr Top Dev Biol. 1998;38:35-80. doi: 10.1016/s0070-2153(08)60244-6. Curr Top Dev Biol. 1998. PMID: 9399076 Review.
Cited by
-
Identification of cis-regulatory modules encoding temporal dynamics during development.BMC Genomics. 2014 Jun 27;15(1):534. doi: 10.1186/1471-2164-15-534. BMC Genomics. 2014. PMID: 24972496 Free PMC article.
-
Extrapolating histone marks across developmental stages, tissues, and species: an enhancer prediction case study.BMC Genomics. 2015 Feb 21;16(1):104. doi: 10.1186/s12864-015-1264-3. BMC Genomics. 2015. PMID: 25765133 Free PMC article.
-
LMethyR-SVM: Predict Human Enhancers Using Low Methylated Regions based on Weighted Support Vector Machines.PLoS One. 2016 Sep 23;11(9):e0163491. doi: 10.1371/journal.pone.0163491. eCollection 2016. PLoS One. 2016. PMID: 27662487 Free PMC article.
-
c-JUN is a barrier in hESC to cardiomyocyte transition.Life Sci Alliance. 2023 Aug 21;6(11):e202302121. doi: 10.26508/lsa.202302121. Print 2023 Nov. Life Sci Alliance. 2023. PMID: 37604584 Free PMC article.
-
Three distinct mechanisms, Notch instructive, permissive, and independent, regulate the expression of two different pericardial genes to specify cardiac cell subtypes.PLoS One. 2020 Oct 27;15(10):e0241191. doi: 10.1371/journal.pone.0241191. eCollection 2020. PLoS One. 2020. PMID: 33108408 Free PMC article.
References
-
- Bailey T. L., Gribskov M. (1998). Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14, 48–54 - PubMed
-
- Barolo S., Posakony J. W. (2002). Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 16, 1167–1181 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

