TGFβ regulates epithelial-mesenchymal interactions through WNT signaling activity to control muscle development in the soft palate
- PMID: 24496627
- PMCID: PMC3912833
- DOI: 10.1242/dev.103093
TGFβ regulates epithelial-mesenchymal interactions through WNT signaling activity to control muscle development in the soft palate
Abstract
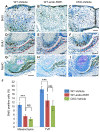
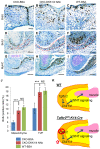
Clefting of the soft palate occurs as a congenital defect in humans and adversely affects the physiological function of the palate. However, the molecular and cellular mechanism of clefting of the soft palate remains unclear because few animal models exhibit an isolated cleft in the soft palate. Using three-dimensional microCT images and histological reconstruction, we found that loss of TGFβ signaling in the palatal epithelium led to soft palate muscle defects in Tgfbr2(fl/fl);K14-Cre mice. Specifically, muscle mass was decreased in the soft palates of Tgfbr2 mutant mice, following defects in cell proliferation and differentiation. Gene expression of Dickkopf (Dkk1 and Dkk4), negative regulators of WNT-β-catenin signaling, is upregulated in the soft palate of Tgfbr2(fl/fl);K14-Cre mice, and WNT-β-catenin signaling is disrupted in the palatal mesenchyme. Importantly, blocking the function of DKK1 and DKK4 rescued the cell proliferation and differentiation defects in the soft palate of Tgfbr2(fl/fl);K14-Cre mice. Thus, our findings indicate that loss of TGFβ signaling in epithelial cells compromises activation of WNT signaling and proper muscle development in the soft palate through tissue-tissue interactions, resulting in a cleft soft palate. This information has important implications for prevention and non-surgical correction of cleft soft palate.
Keywords: Cleft soft palate; Epithelial-mesenchymal interactions; Mouse; TGFβ.
Figures



Similar articles
-
Fibroblast growth factor 9 (FGF9)-pituitary homeobox 2 (PITX2) pathway mediates transforming growth factor β (TGFβ) signaling to regulate cell proliferation in palatal mesenchyme during mouse palatogenesis.J Biol Chem. 2012 Jan 20;287(4):2353-63. doi: 10.1074/jbc.M111.280974. Epub 2011 Nov 28. J Biol Chem. 2012. PMID: 22123828 Free PMC article.
-
Smad4-Irf6 genetic interaction and TGFβ-mediated IRF6 signaling cascade are crucial for palatal fusion in mice.Development. 2013 Mar;140(6):1220-30. doi: 10.1242/dev.089615. Epub 2013 Feb 13. Development. 2013. PMID: 23406900 Free PMC article.
-
Modulation of lipid metabolic defects rescues cleft palate in Tgfbr2 mutant mice.Hum Mol Genet. 2014 Jan 1;23(1):182-93. doi: 10.1093/hmg/ddt410. Epub 2013 Aug 23. Hum Mol Genet. 2014. PMID: 23975680 Free PMC article.
-
Regulatory Mechanisms of Soft Palate Development and Malformations.J Dent Res. 2019 Aug;98(9):959-967. doi: 10.1177/0022034519851786. Epub 2019 May 31. J Dent Res. 2019. PMID: 31150594 Free PMC article. Review.
-
Transforming growth factor beta (TGFbeta) signalling in palatal growth, apoptosis and epithelial mesenchymal transformation (EMT).Arch Oral Biol. 2004 Sep;49(9):675-89. doi: 10.1016/j.archoralbio.2004.05.007. Arch Oral Biol. 2004. PMID: 15275855 Review.
Cited by
-
The Dlx5-FGF10 signaling cascade controls cranial neural crest and myoblast interaction during oropharyngeal patterning and development.Development. 2017 Nov 1;144(21):4037-4045. doi: 10.1242/dev.155176. Epub 2017 Oct 5. Development. 2017. PMID: 28982687 Free PMC article.
-
Chemical-Induced Cleft Palate Is Caused and Rescued by Pharmacological Modulation of the Canonical Wnt Signaling Pathway in a Zebrafish Model.Front Cell Dev Biol. 2020 Dec 14;8:592967. doi: 10.3389/fcell.2020.592967. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33381503 Free PMC article.
-
Genetics and signaling mechanisms of orofacial clefts.Birth Defects Res. 2020 Nov;112(19):1588-1634. doi: 10.1002/bdr2.1754. Epub 2020 Jul 15. Birth Defects Res. 2020. PMID: 32666711 Free PMC article. Review.
-
TGF-β Signaling and the Epithelial-Mesenchymal Transition during Palatal Fusion.Int J Mol Sci. 2018 Nov 19;19(11):3638. doi: 10.3390/ijms19113638. Int J Mol Sci. 2018. PMID: 30463190 Free PMC article. Review.
-
A Comprehensive Study of Soft Palate Development in Mice.PLoS One. 2015 Dec 15;10(12):e0145018. doi: 10.1371/journal.pone.0145018. eCollection 2015. PLoS One. 2015. PMID: 26671681 Free PMC article.
References
-
- Back G. W., Nadig S., Uppal S., Coatesworth A. P. (2004). Why do we have a uvula?: literature review and a new theory. Clin. Otolaryngol. Allied Sci. 29, 689–693 - PubMed
-
- Brault V., Moore R., Kutsch S., Ishibashi M., Rowitch D. H., McMahon A. P., Sommer L., Boussadia O., Kemler R. (2001). Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128, 1253–1264 - PubMed
-
- Chai Y., Maxson R. E., Jr (2006). Recent advances in craniofacial morphogenesis. Dev. Dyn. 235, 2353–2375 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

