Phosphoregulation of Nap1 plays a role in septin ring dynamics and morphogenesis in Candida albicans
- PMID: 24496790
- PMCID: PMC3950511
- DOI: 10.1128/mBio.00915-13
Phosphoregulation of Nap1 plays a role in septin ring dynamics and morphogenesis in Candida albicans
Abstract
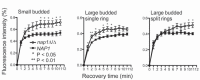
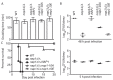
Nap1 has long been identified as a potential septin regulator in yeasts. However, its function and regulation remain poorly defined. Here, we report functional characterization of Nap1 in the human-pathogenic fungus Candida albicans. We find that deletion of NAP1 causes constitutive filamentous growth and changes of septin dynamics. We present evidence that Nap1's cellular localization and function are regulated by phosphorylation. Phos-tag gel electrophoresis revealed that Nap1 phosphorylation is cell cycle dependent, exhibiting the lowest level around the time of bud emergence. Mass spectrometry identified 10 phosphoserine and phosphothreonine residues in a cluster near the N terminus, and mutation of these residues affected Nap1's localization to the septin ring and cellular function. Nap1 phosphorylation involves two septin ring-associated kinases, Cla4 and Gin4, and its dephosphorylation occurs at the septin ring in a manner dependent on the phosphatases PP2A and Cdc14. Furthermore, the nap1Δ/Δ mutant and alleles carrying mutations of the phosphorylation sites exhibited greatly reduced virulence in a mouse model of systemic candidiasis. Together, our findings not only provide new mechanistic insights into Nap1's function and regulation but also suggest the potential to target Nap1 in future therapeutic design.
Importance: Septins are conserved filament-forming GTPases involved in a wide range of cellular events, such as cytokinesis, exocytosis, and morphogenesis. In Candida albicans, the most prevalent human fungal pathogen, septin functions are indispensable for its virulence. However, the molecular mechanisms by which septin structures are regulated are poorly understood. In this study, we deleted NAP1, a gene encoding a putative septin regulator, in C. albicans and found that cells lacking NAP1 showed abnormalities in morphology, invasive growth, and septin ring dynamics. We identified a conserved N-terminal phosphorylation cluster on Nap1 and demonstrated that phosphorylation at these sites regulates Nap1 localization and function. Importantly, deletion of NAP1 or mutation in the N-terminal phosphorylation cluster strongly reduced the virulence of C. albicans in a mouse model of systemic infection. Thus, this study not only provides mechanistic insights into septin regulation but also suggests Nap1 as a potential antifungal target.
Figures
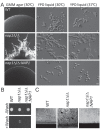
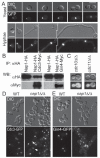

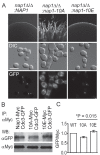
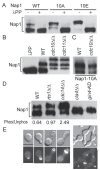

Similar articles
-
Tpd3-Pph21 phosphatase plays a direct role in Sep7 dephosphorylation in Candida albicans.Mol Microbiol. 2016 Jul;101(1):109-21. doi: 10.1111/mmi.13376. Epub 2016 Apr 20. Mol Microbiol. 2016. PMID: 26991697
-
CDK regulates septin organization through cell-cycle-dependent phosphorylation of the Nim1-related kinase Gin4.J Cell Sci. 2012 May 15;125(Pt 10):2533-43. doi: 10.1242/jcs.104497. Epub 2012 Feb 24. J Cell Sci. 2012. PMID: 22366454
-
Rsr1 Palmitoylation and GTPase Activity Status Differentially Coordinate Nuclear, Septin, and Vacuole Dynamics in Candida albicans.mBio. 2020 Oct 13;11(5):e01666-20. doi: 10.1128/mBio.01666-20. mBio. 2020. PMID: 33051364 Free PMC article.
-
Fungal pathogens are platforms for discovering novel and conserved septin properties.Curr Opin Microbiol. 2014 Aug;20:42-8. doi: 10.1016/j.mib.2014.04.004. Epub 2014 May 28. Curr Opin Microbiol. 2014. PMID: 24879478 Free PMC article. Review.
-
Messenger RNA transport in the opportunistic fungal pathogen Candida albicans.Curr Genet. 2017 Dec;63(6):989-995. doi: 10.1007/s00294-017-0707-6. Epub 2017 May 16. Curr Genet. 2017. PMID: 28512683 Free PMC article. Review.
Cited by
-
Novel mechanism coupling cyclic AMP-protein kinase A signaling and golgi trafficking via Gyp1 phosphorylation in polarized growth.Eukaryot Cell. 2014 Dec;13(12):1548-56. doi: 10.1128/EC.00231-14. Epub 2014 Oct 17. Eukaryot Cell. 2014. PMID: 25326521 Free PMC article.
-
Toward a systems-level view of dynamic phosphorylation networks.Front Genet. 2014 Aug 15;5:263. doi: 10.3389/fgene.2014.00263. eCollection 2014. Front Genet. 2014. PMID: 25177341 Free PMC article. Review.
-
From Jekyll to Hyde: The Yeast-Hyphal Transition of Candida albicans.Pathogens. 2021 Jul 7;10(7):859. doi: 10.3390/pathogens10070859. Pathogens. 2021. PMID: 34358008 Free PMC article. Review.
-
A Gin4-Like Protein Kinase GIL1 Involvement in Hyphal Growth, Asexual Development, and Pathogenesis in Fusarium graminearum.Int J Mol Sci. 2017 Feb 16;18(2):424. doi: 10.3390/ijms18020424. Int J Mol Sci. 2017. PMID: 28212314 Free PMC article.
-
A representative of arylcyanomethylenequinone oximes effectively inhibits growth and formation of hyphae in Candida albicans and influences the activity of protein kinases in vitro.Saudi Pharm J. 2018 Feb;26(2):244-252. doi: 10.1016/j.jsps.2017.12.004. Epub 2017 Dec 5. Saudi Pharm J. 2018. PMID: 30166923 Free PMC article.
References
-
- Bertin A, McMurray MA, Grob P, Park SS, Garcia G, III, Patanwala I, Ng HL, Alber T, Thorner J, Nogales E. 2008. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc. Natl. Acad. Sci. U. S. A. 105:8274–8279. 10.1073/pnas.0803330105 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
