Deep conservation of human protein tandem repeats within the eukaryotes
- PMID: 24497029
- PMCID: PMC3995336
- DOI: 10.1093/molbev/msu062
Deep conservation of human protein tandem repeats within the eukaryotes
Abstract
Tandem repeats (TRs) are a major element of protein sequences in all domains of life. They are particularly abundant in mammals, where by conservative estimates one in three proteins contain a TR. High generation-scale duplication and deletion rates were reported for nucleic TR units. However, it is not known whether protein TR units can also be frequently lost or gained providing a source of variation for rapid adaptation of protein function, or alternatively, tend to have conserved TR unit configurations over long evolutionary times. To obtain a systematic picture, we performed a proteome-wide analysis of the mode of evolution for human protein TRs. For this purpose, we propose a novel method for the detection of orthologous TRs based on circular profile hidden Markov models. For all detected TRs, we reconstructed bispecies TR unit phylogenies across 61 eukaryotes ranging from human to yeast. Moreover, we performed additional analyses to correlate functional and structural annotations of human TRs with their mode of evolution. Surprisingly, we find that the vast majority of human TRs are ancient, with TR unit number and order preserved intact since distant speciation events. For example, ≥ 61% of all human TRs have been strongly conserved at least since the root of all mammals, approximately 300 Ma. Further, we find no human protein TR that shows evidence for strong recent duplications and deletions. The results are in contrast to the high generation-scale mutability of nucleic TRs. Presumably, most protein TRs fold into stable and conserved structures that are indispensable for the function of the TR-containing protein. All of our data and results are available for download from http://www.atgc-montpellier.fr/TRE.
Keywords: conservation; phylogenetic analysis; protein evolution; tandem repeats.
Figures
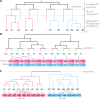
 th TR unit in A is the closest to the
th TR unit in A is the closest to the  th unit in B. Subsequent TR unit duplications and losses diminish the conservation of the TR between species A and B. Without point mutations, the more TR unit losses or gains occur, the more TR units begin to cluster by sequence similarity within the same species. (B) The bi-species TR unit phylogeny of a perfectly conserved WD repeat (PF00400) in the human TORC subunit ENSP00000457870 and its yeast ortholog YNL006W. The TR units are indexed by their order along the protein sequence. The depicted phylogeny allows to reconstruct ancient TR unit duplications leading to the currently observed TR regions in fungi and animals before their divergence ∼0.6–1.6 byr ago (Taylor and Berbee 2007). (C) The bi-species TR unit phylogeny of a perfectly separated TR in the human NAC-alpha domain-containing protein 1 ENSP00000420477 and its mouse ortholog ENSMUSP00000049490. The ancestral protein presumably contained a TR region with multiple repeat units. Yet, the TR region cannot be reconstructed due to the fast succession of TR unit gains/losses in at least one of the lineages.
th unit in B. Subsequent TR unit duplications and losses diminish the conservation of the TR between species A and B. Without point mutations, the more TR unit losses or gains occur, the more TR units begin to cluster by sequence similarity within the same species. (B) The bi-species TR unit phylogeny of a perfectly conserved WD repeat (PF00400) in the human TORC subunit ENSP00000457870 and its yeast ortholog YNL006W. The TR units are indexed by their order along the protein sequence. The depicted phylogeny allows to reconstruct ancient TR unit duplications leading to the currently observed TR regions in fungi and animals before their divergence ∼0.6–1.6 byr ago (Taylor and Berbee 2007). (C) The bi-species TR unit phylogeny of a perfectly separated TR in the human NAC-alpha domain-containing protein 1 ENSP00000420477 and its mouse ortholog ENSMUSP00000049490. The ancestral protein presumably contained a TR region with multiple repeat units. Yet, the TR region cannot be reconstructed due to the fast succession of TR unit gains/losses in at least one of the lineages.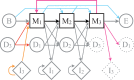

Similar articles
-
The evolution and function of protein tandem repeats in plants.New Phytol. 2015 Apr;206(1):397-410. doi: 10.1111/nph.13184. Epub 2014 Nov 24. New Phytol. 2015. PMID: 25420631
-
Expansion of tandem repeats in sea anemone Nematostella vectensis proteome: A source for gene novelty?BMC Genomics. 2009 Dec 10;10:593. doi: 10.1186/1471-2164-10-593. BMC Genomics. 2009. PMID: 20003297 Free PMC article.
-
Graph-based modeling of tandem repeats improves global multiple sequence alignment.Nucleic Acids Res. 2013 Sep;41(17):e162. doi: 10.1093/nar/gkt628. Epub 2013 Jul 22. Nucleic Acids Res. 2013. PMID: 23877246 Free PMC article.
-
Statistical approaches to detecting and analyzing tandem repeats in genomic sequences.Front Bioeng Biotechnol. 2015 Mar 17;3:31. doi: 10.3389/fbioe.2015.00031. eCollection 2015. Front Bioeng Biotechnol. 2015. PMID: 25853125 Free PMC article. Review.
-
In search of the boundary between repetitive and non-repetitive protein sequences.Biochem Soc Trans. 2015 Oct;43(5):807-11. doi: 10.1042/BST20150073. Biochem Soc Trans. 2015. PMID: 26517886 Review.
Cited by
-
Self-analysis of repeat proteins reveals evolutionarily conserved patterns.BMC Bioinformatics. 2020 May 7;21(1):179. doi: 10.1186/s12859-020-3493-y. BMC Bioinformatics. 2020. PMID: 32381046 Free PMC article.
-
Tandem repeats lead to sequence assembly errors and impose multi-level challenges for genome and protein databases.Nucleic Acids Res. 2019 Dec 2;47(21):10994-11006. doi: 10.1093/nar/gkz841. Nucleic Acids Res. 2019. PMID: 31584084 Free PMC article. Review.
-
Tandem-repeat protein domains across the tree of life.PeerJ. 2015 Jan 13;3:e732. doi: 10.7717/peerj.732. eCollection 2015. PeerJ. 2015. PMID: 25653910 Free PMC article.
-
Accurate contact-based modelling of repeat proteins predicts the structure of new repeats protein families.PLoS Comput Biol. 2021 Apr 15;17(4):e1008798. doi: 10.1371/journal.pcbi.1008798. eCollection 2021 Apr. PLoS Comput Biol. 2021. PMID: 33857128 Free PMC article.
-
Guilt-by-Association - Functional Insights Gained From Studying the LRRK2 Interactome.Front Neurosci. 2020 May 20;14:485. doi: 10.3389/fnins.2020.00485. eCollection 2020. Front Neurosci. 2020. PMID: 32508578 Free PMC article. Review.
References
-
- Abraham A-L, Pothier J, Rocha EPC. Alternative to homo-oligomerisation: the creation of local symmetry in proteins by internal amplification. J Mol Biol. 2009;394:522–534. - PubMed
-
- Angst BD, Marcozzi C, Magee AI. The cadherin superfamily: diversity in form and function. J Cell Sci. 2001;114:629–641. - PubMed
-
- Barford D. The role of multiple sequence repeat motifs in the assembly of multi-protein complexes. In: Carrondo MA, Spadon P, editors. Macromolecular crystallography. Dordrecht (The Netherlands): Springer; 2012. pp. 43–49.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

