Fluoroquinolone-gyrase-DNA complexes: two modes of drug binding
- PMID: 24497635
- PMCID: PMC4007428
- DOI: 10.1074/jbc.M113.529164
Fluoroquinolone-gyrase-DNA complexes: two modes of drug binding
Abstract
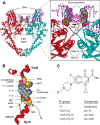
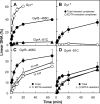
DNA gyrase and topoisomerase IV control bacterial DNA topology by breaking DNA, passing duplex DNA through the break, and then resealing the break. This process is subject to reversible corruption by fluoroquinolones, antibacterials that form drug-enzyme-DNA complexes in which the DNA is broken. The complexes, called cleaved complexes because of the presence of DNA breaks, have been crystallized and found to have the fluoroquinolone C-7 ring system facing the GyrB/ParE subunits. As expected from x-ray crystallography, a thiol-reactive, C-7-modified chloroacetyl derivative of ciprofloxacin (Cip-AcCl) formed cross-linked cleaved complexes with mutant GyrB-Cys(466) gyrase as evidenced by resistance to reversal by both EDTA and thermal treatments. Surprisingly, cross-linking was also readily seen with complexes formed by mutant GyrA-G81C gyrase, thereby revealing a novel drug-gyrase interaction not observed in crystal structures. The cross-link between fluoroquinolone and GyrA-G81C gyrase correlated with exceptional bacteriostatic activity for Cip-AcCl with a quinolone-resistant GyrA-G81C variant of Escherichia coli and its Mycobacterium smegmatis equivalent (GyrA-G89C). Cip-AcCl-mediated, irreversible inhibition of DNA replication provided further evidence for a GyrA-drug cross-link. Collectively these data establish the existence of interactions between the fluoroquinolone C-7 ring and both GyrA and GyrB. Because the GyrA-Gly(81) and GyrB-Glu(466) residues are far apart (17 Å) in the crystal structure of cleaved complexes, two modes of quinolone binding must exist. The presence of two binding modes raises the possibility that multiple quinolone-enzyme-DNA complexes can form, a discovery that opens new avenues for exploring and exploiting relationships between drug structure and activity with type II DNA topoisomerases.
Keywords: Antibiotic Action; Bacteria; Bacterium; Cleaved Complexes; Cross-linking; DNA Topoisomerase; Enzyme Inhibitors; Escherichia coli; Mycobacterium smegmatis; Resistance Mutations.
Figures






References
-
- Champoux J. J. (2001) DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 - PubMed
-
- Wang J. C. (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3, 430–440 - PubMed
-
- Kreuzer K. N., Cozzarelli N. R. (1980) Formation and resolution of DNA catenanes by DNA gyrase. Cell 20, 245–254 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

