Candida albicans utilizes a modified β-oxidation pathway for the degradation of toxic propionyl-CoA
- PMID: 24497638
- PMCID: PMC3961645
- DOI: 10.1074/jbc.M113.517672
Candida albicans utilizes a modified β-oxidation pathway for the degradation of toxic propionyl-CoA
Abstract
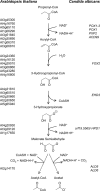
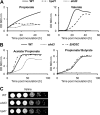
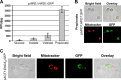
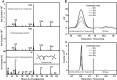
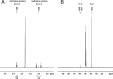
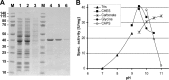
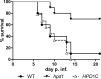
Propionyl-CoA arises as a metabolic intermediate from the degradation of propionate, odd-chain fatty acids, and some amino acids. Thus, pathways for catabolism of this intermediate have evolved in all kingdoms of life, preventing the accumulation of toxic propionyl-CoA concentrations. Previous studies have shown that fungi generally use the methyl citrate cycle for propionyl-CoA degradation. Here, we show that this is not the case for the pathogenic fungus Candida albicans despite its ability to use propionate and valerate as carbon sources. Comparative proteome analyses suggested the presence of a modified β-oxidation pathway with the key intermediate 3-hydroxypropionate. Gene deletion analyses confirmed that the enoyl-CoA hydratase/dehydrogenase Fox2p, the putative 3-hydroxypropionyl-CoA hydrolase Ehd3p, the 3-hydroxypropionate dehydrogenase Hpd1p, and the putative malonate semialdehyde dehydrogenase Ald6p essentially contribute to propionyl-CoA degradation and its conversion to acetyl-CoA. The function of Hpd1p was further supported by the detection of accumulating 3-hydroxypropionate in the hpd1 mutant on propionyl-CoA-generating nutrients. Substrate specificity of Hpd1p was determined from recombinant purified enzyme, which revealed a preference for 3-hydroxypropionate, although serine and 3-hydroxyisobutyrate could also serve as substrates. Finally, virulence studies in a murine sepsis model revealed attenuated virulence of the hpd1 mutant, which indicates generation of propionyl-CoA from host-provided nutrients during infection.
Keywords: Amino Acid; CUG clade; Candida albicans; Enzyme Kinetics; Fatty Acid Metabolism; Fatty Acid Oxidation; Odd-chain Fatty Acids; Pathogenesis; β-Hydroxypropionate.
Figures


Similar articles
-
Peroxisomal fatty acid beta-oxidation is not essential for virulence of Candida albicans.Eukaryot Cell. 2006 Nov;5(11):1847-56. doi: 10.1128/EC.00093-06. Epub 2006 Sep 8. Eukaryot Cell. 2006. PMID: 16963628 Free PMC article.
-
3-hydroxypropionyl-coenzyme A dehydratase and acryloyl-coenzyme A reductase, enzymes of the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle in the Sulfolobales.J Bacteriol. 2009 Jul;191(14):4572-81. doi: 10.1128/JB.00068-09. Epub 2009 May 8. J Bacteriol. 2009. PMID: 19429610 Free PMC article.
-
Role of acetyl coenzyme A synthesis and breakdown in alternative carbon source utilization in Candida albicans.Eukaryot Cell. 2008 Oct;7(10):1733-41. doi: 10.1128/EC.00253-08. Epub 2008 Aug 8. Eukaryot Cell. 2008. PMID: 18689527 Free PMC article.
-
Peroxisomal lipid degradation via beta- and alpha-oxidation in mammals.Cell Biochem Biophys. 2000;32 Spring:73-87. doi: 10.1385/cbb:32:1-3:73. Cell Biochem Biophys. 2000. PMID: 11330072 Review.
-
Intracellular acetyl unit transport in fungal carbon metabolism.Eukaryot Cell. 2010 Dec;9(12):1809-15. doi: 10.1128/EC.00172-10. Epub 2010 Oct 1. Eukaryot Cell. 2010. PMID: 20889721 Free PMC article. Review.
Cited by
-
Inter-relations between 3-hydroxypropionate and propionate metabolism in rat liver: relevance to disorders of propionyl-CoA metabolism.Am J Physiol Endocrinol Metab. 2017 Oct 1;313(4):E413-E428. doi: 10.1152/ajpendo.00105.2017. Epub 2017 Jun 20. Am J Physiol Endocrinol Metab. 2017. PMID: 28634175 Free PMC article.
-
Transcriptome Analyses of Candida albicans Biofilms, Exposed to Arachidonic Acid and Fluconazole, Indicates Potential Drug Targets.G3 (Bethesda). 2020 Sep 2;10(9):3099-3108. doi: 10.1534/g3.120.401340. G3 (Bethesda). 2020. PMID: 32631950 Free PMC article.
-
A streamlined and predominantly diploid genome in the tiny marine green alga Chloropicon primus.Nat Commun. 2019 Sep 6;10(1):4061. doi: 10.1038/s41467-019-12014-x. Nat Commun. 2019. PMID: 31492891 Free PMC article.
-
An integrated transcriptomic and metabolomic approach to investigate the heterogeneous Candida albicans biofilm phenotype.Biofilm. 2023 Mar 12;5:100112. doi: 10.1016/j.bioflm.2023.100112. eCollection 2023 Dec. Biofilm. 2023. PMID: 36969800 Free PMC article.
-
A Comparative Transcriptome Between Anti-drug Sensitive and Resistant Candida auris in China.Front Microbiol. 2021 Jul 16;12:708009. doi: 10.3389/fmicb.2021.708009. eCollection 2021. Front Microbiol. 2021. PMID: 34354695 Free PMC article.
References
-
- Kim J., Sudbery P. (2011) Candida albicans, a major human fungal pathogen. J. Microbiol. 49, 171–177 - PubMed
-
- Gow N. A., Hube B. (2012) Importance of the Candida albicans cell wall during commensalism and infection. Curr. Opin. Microbiol. 15, 406–412 - PubMed
-
- Brock M. (2009) Fungal metabolism in host niches. Curr. Opin. Microbiol. 12, 371–376 - PubMed
-
- Fleck C. B., Schöbel F., Brock M. (2011) Nutrient acquisition by pathogenic fungi: nutrient availability, pathway regulation, and differences in substrate utilization. Int. J. Med. Microbiol. 301, 400–407 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

