Large inverted duplications in the human genome form via a fold-back mechanism
- PMID: 24497845
- PMCID: PMC3907307
- DOI: 10.1371/journal.pgen.1004139
Large inverted duplications in the human genome form via a fold-back mechanism
Abstract
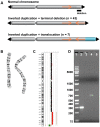
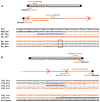
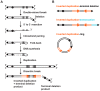
Inverted duplications are a common type of copy number variation (CNV) in germline and somatic genomes. Large duplications that include many genes can lead to both neurodevelopmental phenotypes in children and gene amplifications in tumors. There are several models for inverted duplication formation, most of which include a dicentric chromosome intermediate followed by breakage-fusion-bridge (BFB) cycles, but the mechanisms that give rise to the inverted dicentric chromosome in most inverted duplications remain unknown. Here we have combined high-resolution array CGH, custom sequence capture, next-generation sequencing, and long-range PCR to analyze the breakpoints of 50 nonrecurrent inverted duplications in patients with intellectual disability, autism, and congenital anomalies. For half of the rearrangements in our study, we sequenced at least one breakpoint junction. Sequence analysis of breakpoint junctions reveals a normal-copy disomic spacer between inverted and non-inverted copies of the duplication. Further, short inverted sequences are present at the boundary of the disomic spacer and the inverted duplication. These data support a mechanism of inverted duplication formation whereby a chromosome with a double-strand break intrastrand pairs with itself to form a "fold-back" intermediate that, after DNA replication, produces a dicentric inverted chromosome with a disomic spacer corresponding to the site of the fold-back loop. This process can lead to inverted duplications adjacent to terminal deletions, inverted duplications juxtaposed to translocations, and inverted duplication ring chromosomes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Weleber RG, Verma RS, Kimberling WJ, Fieger HG Jr, lubs HA (1976) Duplication-deficiency of the short arm of chromosome 8 following artificial insemination. Annales de genetique 19: 241–247. - PubMed
-
- Zuffardi O, Bonaglia M, Ciccone R, Giorda R (2009) Inverted duplications deletions: underdiagnosed rearrangements?? Clinical genetics 75: 505–513. - PubMed
-
- Rudd MK (2011) Structural variation in subtelomeres. In: Feuk L, editor. Genomic Structural Variants: Methods and Protocols. New York: Springer Science+Business Media, LLC.
-
- Ballif BC, Yu W, Shaw CA, Kashork CD, Shaffer LG (2003) Monosomy 1p36 breakpoint junctions suggest pre-meiotic breakage-fusion-bridge cycles are involved in generating terminal deletions. Hum Mol Genet 12: 2153–2165. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- SRA/SRP032751
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

