Assessing the spatial dependence of adaptive loci in 43 European and Western Asian goat breeds using AFLP markers
- PMID: 24497965
- PMCID: PMC3907386
- DOI: 10.1371/journal.pone.0086668
Assessing the spatial dependence of adaptive loci in 43 European and Western Asian goat breeds using AFLP markers
Abstract
Background: During the past decades, neutral DNA markers have been extensively employed to study demography, population genetics and structure in livestock, but less interest has been devoted to the evaluation of livestock adaptive potential through the identification of genomic regions likely to be under natural selection.
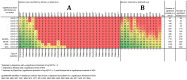
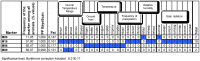
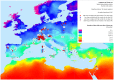
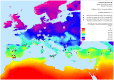
Methodology/principal findings: Landscape genomics can greatly benefit the entire livestock system through the identification of genotypes better adapted to specific or extreme environmental conditions. Therefore we analyzed 101 AFLP markers in 43 European and Western Asian goat breeds both with Matsam software, based on a correlative approach (SAM), and with Mcheza and Bayescan, two FST based software able to detect markers carrying signatures of natural selection. Matsam identified four loci possibly under natural selection--also confirmed by FST-outlier methods--and significantly associated with environmental variables such as diurnal temperature range, frequency of precipitation, relative humidity and solar radiation.
Conclusions/significance: These results show that landscape genomics can provide useful information on the environmental factors affecting the adaptive potential of livestock living in specific climatic conditions. Besides adding conservation value to livestock genetic resources, this knowledge may lead to the development of novel molecular tools useful to preserve the adaptive potential of local breeds during genetic improvement programs, and to increase the adaptability of industrial breeds to changing environments.
Conflict of interest statement
Figures


References
-
- Avise JC (2004) Molecular Markers, Natural History and Evolution. Sunderland Massachusetts: 2nd edn. Sinauer Associates. 684 p.
-
- Cañón J, García D, García-Atance MA, Obexer-Ruff G, Lenstra JA, et al. (2006) Geographical partitioning of goat diversity in Europe and the Middle East. Anim Genet 37: 327–334. - PubMed
-
- SanCristobal M, Chevalet C, Peleman J, Heuven H, Brugmans B, et al. (2006) Genetic diversity in European pigs utilizing amplified fragment length polymorphism markers. Anim Genet 37: 232–238. - PubMed
-
- Luikart G, England PR, Tallmon D, Jordan S, Taberlet P (2003) The power and promise of population genomics: from genotyping to genome typing. Nat Rev Genet 4: 981–994. - PubMed
-
- Vasemägi A, Primmer CR (2005) Challenges for identifying functionally important genetic variation: the promise of combining complementary research strategies. Mol Ecol 14: 3623–3642. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

