Genome-wide detection of copy number variations among diverse horse breeds by array CGH
- PMID: 24497987
- PMCID: PMC3907382
- DOI: 10.1371/journal.pone.0086860
Genome-wide detection of copy number variations among diverse horse breeds by array CGH
Abstract
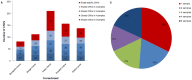
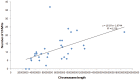
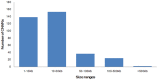
Recent studies have found that copy number variations (CNVs) are widespread in human and animal genomes. CNVs are a significant source of genetic variation, and have been shown to be associated with phenotypic diversity. However, the effect of CNVs on genetic variation in horses is not well understood. In the present study, CNVs in 6 different breeds of mare horses, Mongolia horse, Abaga horse, Hequ horse and Kazakh horse (all plateau breeds) and Debao pony and Thoroughbred, were determined using aCGH. In total, seven hundred CNVs were identified ranging in size from 6.1 Kb to 0.57 Mb across all autosomes, with an average size of 43.08 Kb and a median size of 15.11 Kb. By merging overlapping CNVs, we found a total of three hundred and fifty-three CNV regions (CNVRs). The length of the CNVRs ranged from 6.1 Kb to 1.45 Mb with average and median sizes of 38.49 Kb and 13.1 Kb. Collectively, 13.59 Mb of copy number variation was identified among the horses investigated and accounted for approximately 0.61% of the horse genome sequence. Five hundred and eighteen annotated genes were affected by CNVs, which corresponded to about 2.26% of all horse genes. Through the gene ontology (GO), genetic pathway analysis and comparison of CNV genes among different breeds, we found evidence that CNVs involving 7 genes may be related to the adaptation to severe environment of these plateau horses. This study is the first report of copy number variations in Chinese horses, which indicates that CNVs are ubiquitous in the horse genome and influence many biological processes of the horse. These results will be helpful not only in mapping the horse whole-genome CNVs, but also to further research for the adaption to the high altitude severe environment for plateau horses.
Conflict of interest statement
Figures

Similar articles
-
Inter- and intra-breed genome-wide copy number diversity in a large cohort of European equine breeds.BMC Genomics. 2019 Oct 22;20(1):759. doi: 10.1186/s12864-019-6141-z. BMC Genomics. 2019. PMID: 31640551 Free PMC article.
-
Genome-Wide Detection of Copy Number Variants in Chinese Indigenous Horse Breeds and Verification of CNV-Overlapped Genes Related to Heat Adaptation of the Jinjiang Horse.Genes (Basel). 2022 Mar 28;13(4):603. doi: 10.3390/genes13040603. Genes (Basel). 2022. PMID: 35456409 Free PMC article.
-
Identification of copy number variants in horses.Genome Res. 2012 May;22(5):899-907. doi: 10.1101/gr.128991.111. Epub 2012 Mar 1. Genome Res. 2012. PMID: 22383489 Free PMC article.
-
Advancements in copy number variation screening in herbivorous livestock genomes and their association with phenotypic traits.Front Vet Sci. 2024 Jan 11;10:1334434. doi: 10.3389/fvets.2023.1334434. eCollection 2023. Front Vet Sci. 2024. PMID: 38274664 Free PMC article. Review.
-
Copy number variants and selective sweeps in natural populations of the house mouse (Mus musculus domesticus).Front Genet. 2014 Jun 3;5:153. doi: 10.3389/fgene.2014.00153. eCollection 2014. Front Genet. 2014. PMID: 24917877 Free PMC article. Review.
Cited by
-
Molecular subtyping of ependymoma and prognostic impact of Ki-67.Brain Tumor Pathol. 2022 Jan;39(1):1-13. doi: 10.1007/s10014-021-00417-y. Epub 2021 Nov 23. Brain Tumor Pathol. 2022. PMID: 34812989 Free PMC article.
-
CNVs with adaptive potential in Rangifer tarandus: genome architecture and new annotated assembly.Life Sci Alliance. 2021 Dec 15;5(3):e202101207. doi: 10.26508/lsa.202101207. Print 2022 Mar. Life Sci Alliance. 2021. PMID: 34911809 Free PMC article.
-
Whole genome detection of sequence and structural polymorphism in six diverse horses.PLoS One. 2020 Apr 9;15(4):e0230899. doi: 10.1371/journal.pone.0230899. eCollection 2020. PLoS One. 2020. PMID: 32271776 Free PMC article.
-
Improved de novo genomic assembly for the domestic donkey.Sci Adv. 2018 Apr 4;4(4):eaaq0392. doi: 10.1126/sciadv.aaq0392. eCollection 2018 Apr. Sci Adv. 2018. PMID: 29740610 Free PMC article.
-
Mutation Analysis of Cell-Free DNA and Single Circulating Tumor Cells in Metastatic Breast Cancer Patients with High Circulating Tumor Cell Counts.Clin Cancer Res. 2017 Jan 1;23(1):88-96. doi: 10.1158/1078-0432.CCR-16-0825. Epub 2016 Jun 22. Clin Cancer Res. 2017. PMID: 27334837 Free PMC article.
References
-
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, et al. (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409: 928–933. - PubMed
-
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, et al. (2004) Large-scale copynumber polymorphism in the human genome. Science 305: 525–528. - PubMed
-
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, et al. (2004) Detection of large-scale variation in the human genome. Nat Genet 36: 949–951. - PubMed
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

