Crystal structure of an (R)-selective ω-transaminase from Aspergillus terreus
- PMID: 24498081
- PMCID: PMC3907554
- DOI: 10.1371/journal.pone.0087350
Crystal structure of an (R)-selective ω-transaminase from Aspergillus terreus
Abstract
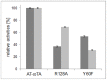
Chiral amines are important building blocks for the synthesis of pharmaceutical products, fine chemicals, and agrochemicals. ω-Transaminases are able to directly synthesize enantiopure chiral amines by catalysing the transfer of an amino group from a primary amino donor to a carbonyl acceptor with pyridoxal 5'-phosphate (PLP) as cofactor. In nature, (S)-selective amine transaminases are more abundant than the (R)-selective enzymes, and therefore more information concerning their structures is available. Here, we present the crystal structure of an (R)-ω-transaminase from Aspergillus terreus determined by X-ray crystallography at a resolution of 1.6 Å. The structure of the protein is a homodimer that displays the typical class IV fold of PLP-dependent aminotransferases. The PLP-cofactor observed in the structure is present in two states (i) covalently bound to the active site lysine (the internal aldimine form) and (ii) as substrate/product adduct (the external aldimine form) and free lysine. Docking studies revealed that (R)-transaminases follow a dual binding mode, in which the large binding pocket can harbour the bulky substituent of the amine or ketone substrate and the α-carboxylate of pyruvate or amino acids, and the small binding pocket accommodates the smaller substituent.
Conflict of interest statement
Figures


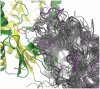
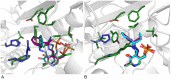


References
-
- Breuer M, Ditrich K, Habicher T, Hauer B, Kesseler M, et al. (2004) Industrial methods for the production of optically active intermediates. Angew Chem Int Ed 43: 788–824. - PubMed
-
- Höhne M, Bornscheuer U (2009) Biocatalytic routes to optically active amines. ChemCatChem 1: 42–51.
-
- Koszelewski D, Göritzer M, Clay D, Seisser B, Kroutil W (2010) Synthesis of optically active amines employing recombinant ω-transaminases in E. coli cells. ChemCatChem 2: 73–77.
-
- Carr R, Alexeeva M, Dawson M, Gotor-Fernandez V, Humphrey C, et al. (2005) Directed evolution of an amine oxidase for the preparative deracemisation of cyclic secondary amines. ChemBioChem 6: 637–639. - PubMed
-
- Ismail H, Lau R, van Rantwijk F, Sheldon R (2008) Fully enzymatic resolution of chiral amines: acylation and deacylation in the presence of Candida antarctica lipase B. Adv Synth Catal. 350: 1511–1516.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

