Regulation of tissue LC-PUFA contents, Δ6 fatty acyl desaturase (FADS2) gene expression and the methylation of the putative FADS2 gene promoter by different dietary fatty acid profiles in Japanese seabass (Lateolabrax japonicus)
- PMID: 24498178
- PMCID: PMC3909213
- DOI: 10.1371/journal.pone.0087726
Regulation of tissue LC-PUFA contents, Δ6 fatty acyl desaturase (FADS2) gene expression and the methylation of the putative FADS2 gene promoter by different dietary fatty acid profiles in Japanese seabass (Lateolabrax japonicus)
Abstract
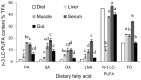
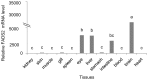
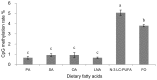
The present study was conducted to evaluate the influences of different dietary fatty acid profiles on the tissue content and biosynthesis of LC-PUFA in a euryhaline species Japanese seabass reared in seawater. Six diets were prepared, each with a characteristic fatty acid: Diet PA: Palmitic acid (C16:0); Diet SA: Stearic acid (C18:0); Diet OA: Oleic acid (C18:1n-9); Diet LNA: α-linolenic acid (C18:3n-3); Diet N-3 LC-PUFA: n-3 LC-PUFA (DHA+EPA); Diet FO: the fish oil control. A 10-week feeding trial was conducted using juvenile fish (29.53 ± 0.86 g). The results showed that Japanese seabass had limited capacity to synthesize LC-PUFA and fish fed PA, SA, OA and LNA showed significantly lower tissue n-3 LC-PUFA contents compared to fish fed N-3 LC-PUFA and FO. The putative gene promoter and full-length cDNA of FADS2 was cloned and characterized. The protein sequence was confirmed to be homologous to FADS2s of marine teleosts and possessed all the characteristic features of microsomal fatty acid desaturases. The FADS2 transcript levels in liver of fish fed N-3 LC-PUFA and FO were significantly lower than those in fish fed other diets except LNA while Diet PA significantly up-regulated the FADS2 gene expression compared to Diet LNA, N-3 LC-PUFA and FO. Inversely, fish fed N-3 LC-PUFA and FO showed significantly higher promoter methylation rates of FADS2 gene compared to fish fed the LC-PUFA deficient diets. These results suggested that Japanese seabass had low LC-PUFA synthesis capacity and LC-PUFA deficient diets caused significantly reduced tissue n-3 LC-PUFA contents. The liver gene expression of FADS2 was up-regulated in groups enriched in C16:0, C18:0 and C18:1n-9 respectively but not in the group enriched in C18:3n-3 compared to groups with high n-3 LC-PUFA contents. The FADS2 gene expression regulated by dietary fatty acids was significantly negatively correlated with the methylation rate of putative FADS2 gene promoter.
Conflict of interest statement
Figures




References
-
- Howe PR, Downing JA, Grenyer BF, Grigonis-Deane EM, Bryden WL (2002) Tuna fishmeal as a source of DHA for n−3 PUFA enrichment of pork, chicken, and eggs. Lipids 37: 1067–1076. - PubMed
-
- Sioutis S, Coates AM, Buckley JD, Murphy TW, Channon HA, et al. (2008) N−3 enrichment of pork with fishmeal: Effects on production and consumer acceptability. Eur J Lipid Sci Technol 110: 701–706.
-
- Tacon AG, Metian M (2008) Global overview on the use of fish meal and fish oil in industrially compounded aquafeeds: Trends and future prospects. Aquaculture 285: 146–158.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

