MT-CYB mutations in hypertrophic cardiomyopathy
- PMID: 24498601
- PMCID: PMC3893158
- DOI: 10.1002/mgg3.5
MT-CYB mutations in hypertrophic cardiomyopathy
Erratum in
- Mol Genet Genomic Med. 2013 Sep;1(3):187
Abstract
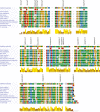
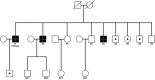
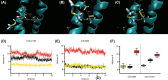
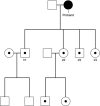
Mitochondrial dysfunction is a characteristic of heart failure. Mutations in mitochondrial DNA, particularly in MT-CYB coding for cytochrome B in complex III (CIII), have been associated with isolated hypertrophic cardiomyopathy (HCM). We hypothesized that MT-CYB mutations might play an important causal or modifying role in HCM. The MT-CYB gene was sequenced from DNA isolated from blood from 91 Danish HCM probands. Nonsynonymous variants were analyzed by bioinformatics, molecular modeling and simulation. Two germline-inherited, putative disease-causing, nonsynonymous variants: m.15024G>A; p.C93Y and m.15482T>C; p.S246P were identified. Modeling showed that the p.C93Y mutation leads to disruption of the tertiary structure of Cytb by helix displacement, interfering with protein-heme interaction. The p.S246P mutation induces a diproline structure, which alters local secondary structure and induces a kink in the protein backbone, interfering with macromolecular interactions. These molecular effects are compatible with a leaky phenotype, that is, limited but progressive mitochondrial dysfunction. In conclusion, we find that rare, putative leaky mtDNA variants in MT-CYB can be identified in a cohort of HCM patients. We propose that further patients with HCM should be examined for mutations in MT-CYB in order to clarify the role of these variants.
Keywords: Cardiomyopathy; DNA sequencing; genetic disorders; hypertrophy; mitochondria.
Figures
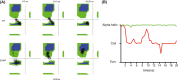

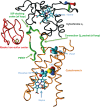
References
-
- Andersen PS, Havndrup O, Hougs L, Sorensen KM, Jensen M, Larsen LA, et al. Diagnostic yield, interpretation, and clinical utility of mutation screening of sarcomere encoding genes in Danish hypertrophic cardiomyopathy patients and relatives. Hum. Mutat. 2009;30:363–370. - PubMed
-
- Andreu AL, Checcarelli N, Iwata S, Shanske S, DiMauro S. A missense mutation in the mitochondrial cytochrome b gene in a revisited case with histiocytoid cardiomyopathy. Pediatr. Res. 2000;48:311–314. - PubMed
-
- Ballana E, Govea N, Garcia R, de Cid C, Arribas C, Rosell J, et al. Detection of unrecognized low-level mtDNA heteroplasmy may explain the variable phenotypic expressivity of apparently homoplasmic mtDNA mutations. Hum. Mutat. 2008;29:248–257. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

