Antisense suppression of donor splice site mutations in the dystrophin gene transcript
- PMID: 24498612
- PMCID: PMC3865583
- DOI: 10.1002/mgg3.19
Antisense suppression of donor splice site mutations in the dystrophin gene transcript
Abstract
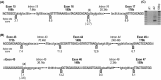
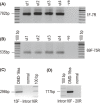
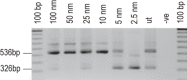
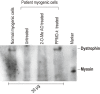
We describe two donor splice site mutations, affecting dystrophin exons 16 and 45 that led to Duchenne muscular dystrophy (DMD), through catastrophic inactivation of the mRNA. These gene lesions unexpectedly resulted in the retention of the downstream introns, thereby increasing the length of the dystrophin mRNA by 20.2 and 36 kb, respectively. Splice-switching antisense oligomers targeted to exon 16 excised this in-frame exon and the following intron from the patient dystrophin transcript very efficiently in vitro, thereby restoring the reading frame and allowing synthesis of near-normal levels of a putatively functional dystrophin isoform. In contrast, targeting splice-switching oligomers to exon 45 in patient cells promoted only modest levels of an out-of-frame dystrophin transcript after transfection at high oligomer concentrations, whereas dual targeting of exons 44 and 45 or 45 and 46 resulted in more efficient exon skipping, with concomitant removal of intron 45. The splice site mutations reported here appear highly amenable to antisense oligomer intervention. We suggest that other splice site mutations may need to be evaluated for oligomer interventions on a case-by-case basis.
Keywords: Duchenne muscular dystrophy; exon skipping; splice mutation; splice-switching oligomer.
Figures

Similar articles
-
Targeted exon skipping to address "leaky" mutations in the dystrophin gene.Mol Ther Nucleic Acids. 2012 Oct 16;1(10):e48. doi: 10.1038/mtna.2012.40. Mol Ther Nucleic Acids. 2012. PMID: 23344648 Free PMC article.
-
Mismatched single stranded antisense oligonucleotides can induce efficient dystrophin splice switching.BMC Med Genet. 2011 Oct 20;12:141. doi: 10.1186/1471-2350-12-141. BMC Med Genet. 2011. PMID: 22013876 Free PMC article.
-
DMD antisense oligonucleotide mediated exon skipping efficiency correlates with flanking intron retention time and target position within the exon.RNA Biol. 2023 Jan;20(1):693-702. doi: 10.1080/15476286.2023.2254041. RNA Biol. 2023. PMID: 37667454 Free PMC article.
-
Splice modification to restore functional dystrophin synthesis in Duchenne muscular dystrophy.Curr Pharm Des. 2010;16(8):988-1001. doi: 10.2174/138161210790883480. Curr Pharm Des. 2010. PMID: 20041827 Review.
-
Molecular correction of Duchenne muscular dystrophy by splice modulation and gene editing.RNA Biol. 2021 Jul;18(7):1048-1062. doi: 10.1080/15476286.2021.1874161. Epub 2021 Jan 20. RNA Biol. 2021. PMID: 33472516 Free PMC article. Review.
Cited by
-
LOX-1 and Its Splice Variants: A New Challenge for Atherosclerosis and Cancer-Targeted Therapies.Int J Mol Sci. 2017 Jan 29;18(2):290. doi: 10.3390/ijms18020290. Int J Mol Sci. 2017. PMID: 28146073 Free PMC article. Review.
-
Systematic analysis of the effects of splicing on the diversity of post-translational modifications in protein isoforms using PTM-POSE.Cell Syst. 2025 Jul 16;16(7):101318. doi: 10.1016/j.cels.2025.101318. Epub 2025 Jun 12. Cell Syst. 2025. PMID: 40513562
-
Pseudoexon activation increases phenotype severity in a Becker muscular dystrophy patient.Mol Genet Genomic Med. 2015 Jul;3(4):320-6. doi: 10.1002/mgg3.144. Epub 2015 Apr 15. Mol Genet Genomic Med. 2015. PMID: 26247048 Free PMC article.
-
Artificial Intelligence, Healthcare, Clinical Genomics, and Pharmacogenomics Approaches in Precision Medicine.Front Genet. 2022 Jul 6;13:929736. doi: 10.3389/fgene.2022.929736. eCollection 2022. Front Genet. 2022. PMID: 35873469 Free PMC article. Review.
-
The identification of protein and RNA interactors of the splicing factor Caper in the adult Drosophila nervous system.Front Mol Neurosci. 2023 Jun 23;16:1114857. doi: 10.3389/fnmol.2023.1114857. eCollection 2023. Front Mol Neurosci. 2023. PMID: 37435576 Free PMC article.
References
-
- Aartsma-Rus A, Fokkema I, Verschuuren J, Ginjaar I, van Deutekom J, van Ommen GJ, et al. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum. Mutat. 2009;30:293–299. - PubMed
-
- Adachi K, Takeshima Y, Wada H, Yagi M, Nakamura H, Matsuo M. Heterogous dystrophin mRNA produced by a novel splice acceptor site mutation in intermediate dystrophinopathy. Pediatr. Res. 2003;53:125–131. - PubMed
-
- Arechavala-Gomeza V, Graham IR, Popplewell LJ, Adams AM, Aartsma-Rus A, Kinali M, et al. Comparative analysis of antisense oligonucleotide sequences for targeted skipping of exon 51 during dystrophin Pre-MRNA splicing in human muscle. Hum. Gene Ther. 2007;18:798–810. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

