Bacterial insertion sequences: their genomic impact and diversity
- PMID: 24499397
- PMCID: PMC7190074
- DOI: 10.1111/1574-6976.12067
Bacterial insertion sequences: their genomic impact and diversity
Abstract
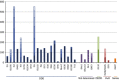
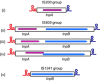
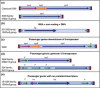
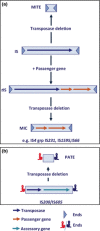
Insertion sequences (ISs), arguably the smallest and most numerous autonomous transposable elements (TEs), are important players in shaping their host genomes. This review focuses on prokaryotic ISs. We discuss IS distribution and impact on genome evolution. We also examine their effects on gene expression, especially their role in activating neighbouring genes, a phenomenon of particular importance in the recent upsurge of bacterial antibiotic resistance. We explain how ISs are identified and classified into families by a combination of characteristics including their transposases (Tpases), their overall genetic organisation and the accessory genes which some ISs carry. We then describe the organisation of autonomous and nonautonomous IS-related elements. This is used to illustrate the growing recognition that the boundaries between different types of mobile element are becoming increasingly difficult to define as more are being identified. We review the known Tpase types, their different catalytic activities used in cleaving and rejoining DNA strands during transposition, their organisation into functional domains and the role of this in regulation. Finally, we consider examples of prokaryotic IS domestication. In a more speculative section, we discuss the necessity of constructing more quantitative dynamic models to fully appreciate the continuing impact of TEs on prokaryotic populations.
Keywords: diversity; evolution; genome; insertion sequence; mechanism.
© 2014 Federation of European Microbiological Societies. Published by John Wiley & Sons Ltd. All rights reserved.
Figures

References
-
- Aguena M, Ferreira GM & Spira B (2009) Stability of the pstS transcript of Escherichia coli . Arch Microbiol 191: 105–112. - PubMed
-
- Allewalt JP, Bateson MM, Revsbech NP, Slack K & Ward DM (2006) Effect of temperature and light on growth of and photosynthesis by Synechococcus isolates typical of those predominating in the octopus spring microbial mat community of Yellowstone National Park. Appl Environ Microbiol 72: 544–550. - PMC - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

