Life in an unusual intracellular niche: a bacterial symbiont infecting the nucleus of amoebae
- PMID: 24500618
- PMCID: PMC4817620
- DOI: 10.1038/ismej.2014.5
Life in an unusual intracellular niche: a bacterial symbiont infecting the nucleus of amoebae
Abstract
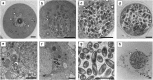
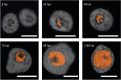
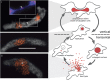
Amoebae serve as hosts for various intracellular bacteria, including human pathogens. These microbes are able to overcome amoebal defense mechanisms and successfully establish a niche for replication, which is usually the cytoplasm. Here, we report on the discovery of a bacterial symbiont that is located inside the nucleus of its Hartmannella sp. host. This symbiont, tentatively named 'Candidatus Nucleicultrix amoebiphila', is only moderately related to known bacteria (∼90% 16S and 23S rRNA sequence similarity) and member of a novel clade of protist symbionts affiliated with the Rickettsiales and Rhodospirillales. Screening of 16S rRNA amplicon data sets revealed a broad distribution of these bacteria in freshwater and soil habitats. 'Candidatus Nucleicultrix amoebiphila' traffics within 6 h post infection to the host nucleus. Maximum infection levels are reached after 96-120 h, at which time point the nucleus is pronouncedly enlarged and filled with bacteria. Transmission of the symbionts occurs vertically upon host cell division but may also occur horizontally through host cell lysis. Although we observed no impact on the fitness of the original Hartmannella sp. host, the bacteria are rather lytic for Acanthamoeba castellanii. Intranuclear symbiosis is an exceptional phenomenon, and amoebae represent an ideal model system to further investigate evolution and underlying molecular mechanisms of these unique microbial associations.
Figures



Similar articles
-
A Rickettsiales symbiont of amoebae with ancient features.Environ Microbiol. 2016 Sep;18(8):2326-42. doi: 10.1111/1462-2920.12881. Epub 2015 Jun 5. Environ Microbiol. 2016. PMID: 25908022
-
Disentangling the Taxonomy of Rickettsiales and Description of Two Novel Symbionts ("Candidatus Bealeia paramacronuclearis" and "Candidatus Fokinia cryptica") Sharing the Cytoplasm of the Ciliate Protist Paramecium biaurelia.Appl Environ Microbiol. 2016 Nov 21;82(24):7236-7247. doi: 10.1128/AEM.02284-16. Print 2016 Dec 15. Appl Environ Microbiol. 2016. PMID: 27742680 Free PMC article.
-
Comparative Genomic Analysis of Acanthamoeba Endosymbionts Highlights the Role of Amoebae as a "Melting Pot" Shaping the Rickettsiales Evolution.Genome Biol Evol. 2017 Nov 1;9(11):3214-3224. doi: 10.1093/gbe/evx246. Genome Biol Evol. 2017. PMID: 29177480 Free PMC article.
-
Bacterial endosymbionts of free-living amoebae.J Eukaryot Microbiol. 2004 Sep-Oct;51(5):509-14. doi: 10.1111/j.1550-7408.2004.tb00278.x. J Eukaryot Microbiol. 2004. PMID: 15537084 Review.
-
Intranuclear bacteria: inside the cellular control center of eukaryotes.Trends Cell Biol. 2015 Jun;25(6):339-46. doi: 10.1016/j.tcb.2015.01.002. Epub 2015 Feb 10. Trends Cell Biol. 2015. PMID: 25680230 Review.
Cited by
-
Vermamoeba vermiformis: a Free-Living Amoeba of Interest.Microb Ecol. 2018 Nov;76(4):991-1001. doi: 10.1007/s00248-018-1199-8. Epub 2018 May 8. Microb Ecol. 2018. PMID: 29737382 Review.
-
Commensals Serve as Natural Barriers to Mammalian Cells during Acanthamoeba castellanii Invasion.Microbiol Spectr. 2021 Dec 22;9(3):e0051221. doi: 10.1128/Spectrum.00512-21. Epub 2021 Dec 22. Microbiol Spectr. 2021. PMID: 34935418 Free PMC article.
-
"Candidatus Intestinibacterium parameciiphilum"-member of the "Candidatus Paracaedibacteraceae" family (Alphaproteobacteria, Holosporales) inhabiting the ciliated protist Paramecium.Int Microbiol. 2024 Jun;27(3):659-671. doi: 10.1007/s10123-023-00414-5. Epub 2023 Aug 24. Int Microbiol. 2024. PMID: 37615902 Free PMC article.
-
An Evolutionary-Focused Review of the Holosporales (Alphaproteobacteria): Diversity, Host Interactions, and Taxonomic Re-ranking as Holosporineae Subord. Nov.Microb Ecol. 2025 Mar 14;88(1):15. doi: 10.1007/s00248-025-02509-0. Microb Ecol. 2025. PMID: 40085262 Free PMC article. Review.
-
'Candidatus Cochliophilus cryoturris' (Coxiellaceae), a symbiont of the testate amoeba Cochliopodium minus.Sci Rep. 2017 Jun 13;7(1):3394. doi: 10.1038/s41598-017-03642-8. Sci Rep. 2017. PMID: 28611430 Free PMC article.
References
-
- Alverca E, Biegala I, Kennaway G. (2002). In situ identification and localization of bacteria associated with Gyrodinium instriatum (Gymnodiniales, Dinophyceae) by electron and confocal microscopy. Eur J Phycol 37: 37–41.
-
- Barker J, Brown M. (1994). Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology 140: 1253–1259. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

