Centromere reference models for human chromosomes X and Y satellite arrays
- PMID: 24501022
- PMCID: PMC3975068
- DOI: 10.1101/gr.159624.113
Centromere reference models for human chromosomes X and Y satellite arrays
Abstract
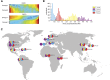
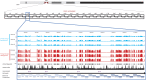
The human genome sequence remains incomplete, with multimegabase-sized gaps representing the endogenous centromeres and other heterochromatic regions. Available sequence-based studies within these sites in the genome have demonstrated a role in centromere function and chromosome pairing, necessary to ensure proper chromosome segregation during cell division. A common genomic feature of these regions is the enrichment of long arrays of near-identical tandem repeats, known as satellite DNAs, which offer a limited number of variant sites to differentiate individual repeat copies across millions of bases. This substantial sequence homogeneity challenges available assembly strategies and, as a result, centromeric regions are omitted from ongoing genomic studies. To address this problem, we utilize monomer sequence and ordering information obtained from whole-genome shotgun reads to model two haploid human satellite arrays on chromosomes X and Y, resulting in an initial characterization of 3.83 Mb of centromeric DNA within an individual genome. To further expand the utility of each centromeric reference sequence model, we evaluate sites within the arrays for short-read mappability and chromosome specificity. Because satellite DNAs evolve in a concerted manner, we use these centromeric assemblies to assess the extent of sequence variation among 366 individuals from distinct human populations. We thus identify two satellite array variants in both X and Y centromeres, as determined by array length and sequence composition. This study provides an initial sequence characterization of a regional centromere and establishes a foundation to extend genomic characterization to these sites as well as to other repeat-rich regions within complex genomes.
Figures


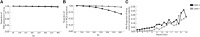
References
-
- Alexandrov IA, Mitkevich SP, Yurov YB 1988. The phylogeny of human chromosome specific α satellites. Chromosoma 96: 443–453 - PubMed
-
- Chang C, Lin C 2011. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol 2: 1–27
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
