Rapid and specific detection, molecular epidemiology, and experimental virulence of the O16 subgroup within Escherichia coli sequence type 131
- PMID: 24501035
- PMCID: PMC3993632
- DOI: 10.1128/JCM.03502-13
Rapid and specific detection, molecular epidemiology, and experimental virulence of the O16 subgroup within Escherichia coli sequence type 131
Abstract
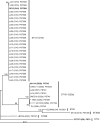
Escherichia coli sequence type 131 (ST131), a widely disseminated multidrug-resistant extraintestinal pathogen, typically exhibits serotype O25b:H4. However, certain ST131 isolates exhibit serotype O16:H5 and derive from a phylogenetic clade that is distinct from the classic O25b:H4 ST131 clade. Both clades are assigned to ST131 by the Achtman multilocus sequence typing (MLST) system and a screening PCR assay that targets ST131-specific sequence polymorphisms in the mdh and gyrB genes. However, they are classified as separate STs by the Pasteur Institute MLST system, and an ST131 PCR method that targets the O25b rfb region and an ST131-specific polymorphism in pabB detects only the O25b-associated clade. Here, we describe a novel PCR-based method that allows for rapid and specific detection of the O16-associated ST131 clade. The clade members uniformly contained allele 41 of fimH (type 1 fimbrial adhesin) and a narrow range of alleles of gyrA and parC (fluoroquinolone target genes). The virulence genotypes of the clade members resembled those of classic O25b:H4 ST131 isolates; representative isolates were variably lethal in a mouse subcutaneous sepsis model. Several pulsotypes spanned multiple sources (adults, children, pets, and human fecal samples) and locales. An analysis of recent clinical E. coli collections showed that the O16 ST131 clade is globally distributed, accounts for 1 to 5% of E. coli isolates overall, and, when compared with other ST131 isolates, it is associated with resistance to ampicillin, gentamicin, and trimethoprim-sulfamethoxazole and with susceptibility to fluoroquinolones and extended-spectrum cephalosporins. Attention to this O16-associated ST131 clade, which is facilitated by our novel PCR-based assay, is warranted in future epidemiological studies of ST131 and, conceivably, in clinical applications.
Figures

References
-
- Colpan A, Johnston B, Porter S, Clabots C, Anway R, Thao L, Kuskowski MA, Tchesnokova V, Sokurenko EV, Johnson JR, VICTORY (Veterans Influence of Clonal Types on Resistance: Year 2011) Investigators 2013. Escherichia coli sequence type 131 (ST131) as an emergent multidrug-resistant pathogen among U.S. veterans. Clin. Infect. Dis. 57:1256–1265. 10.1093/cid/cit503 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

