Universal influenza B virus genomic amplification facilitates sequencing, diagnostics, and reverse genetics
- PMID: 24501036
- PMCID: PMC3993638
- DOI: 10.1128/JCM.03265-13
Universal influenza B virus genomic amplification facilitates sequencing, diagnostics, and reverse genetics
Abstract
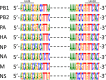
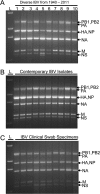
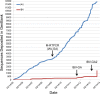
Although human influenza B virus (IBV) is a significant human pathogen, its great genetic diversity has limited our ability to universally amplify the entire genome for subsequent sequencing or vaccine production. The generation of sequence data via next-generation approaches and the rapid cloning of viral genes are critical for basic research, diagnostics, antiviral drugs, and vaccines to combat IBV. To overcome the difficulty of amplifying the diverse and ever-changing IBV genome, we developed and optimized techniques that amplify the complete segmented negative-sense RNA genome from any IBV strain in a single tube/well (IBV genomic amplification [IBV-GA]). Amplicons for >1,000 diverse IBV genomes from different sample types (e.g., clinical specimens) were generated and sequenced using this robust technology. These approaches are sensitive, robust, and sequence independent (i.e., universally amplify past, present, and future IBVs), which facilitates next-generation sequencing and advanced genomic diagnostics. Importantly, special terminal sequences engineered into the optimized IBV-GA2 products also enable ligation-free cloning to rapidly generate reverse-genetics plasmids, which can be used for the rescue of recombinant viruses and/or the creation of vaccine seed stock.
Figures


References
-
- Feng L, Shay DK, Jiang Y, Zhou H, Chen X, Zheng Y, Jiang L, Zhang Q, Lin H, Wang S, Ying Y, Xu Y, Wang N, Feng Z, Viboud C, Yang W, Yu H. 2012. Influenza-associated mortality in temperate and subtropical Chinese cities, 2003–2008. Bull. World Health Organ. 90:279–288B. 10.2471/BLT.11.096958 - DOI - PMC - PubMed
-
- WHO. 2009. Influenza (seasonal) fact sheet no. 211. World Health Organization, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs211/en/
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

