Cell lineage tracing in the developing enteric nervous system: superstars revealed by experiment and simulation
- PMID: 24501272
- PMCID: PMC3928926
- DOI: 10.1098/rsif.2013.0815
Cell lineage tracing in the developing enteric nervous system: superstars revealed by experiment and simulation
Abstract
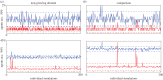
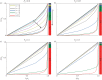
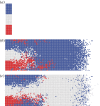
Cell lineage tracing is a powerful tool for understanding how proliferation and differentiation of individual cells contribute to population behaviour. In the developing enteric nervous system (ENS), enteric neural crest (ENC) cells move and undergo massive population expansion by cell division within self-growing mesenchymal tissue. We show that single ENC cells labelled to follow clonality in the intestine reveal extraordinary and unpredictable variation in number and position of descendant cells, even though ENS development is highly predictable at the population level. We use an agent-based model to simulate ENC colonization and obtain agent lineage tracing data, which we analyse using econometric data analysis tools. In all realizations, a small proportion of identical initial agents accounts for a substantial proportion of the total final agent population. We term these individuals superstars. Their existence is consistent across individual realizations and is robust to changes in model parameters. This inequality of outcome is amplified at elevated proliferation rate. The experiments and model suggest that stochastic competition for resources is an important concept when understanding biological processes which feature high levels of cell proliferation. The results have implications for cell-fate processes in the ENS.
Keywords: cell lineage; cell-fate decisions; enteric nervous system; invasion wave.
Figures
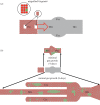

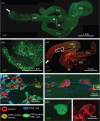

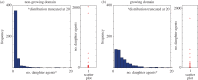

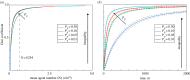


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

