Transgenic mouse lines subdivide external segment of the globus pallidus (GPe) neurons and reveal distinct GPe output pathways
- PMID: 24501350
- PMCID: PMC3913864
- DOI: 10.1523/JNEUROSCI.4646-13.2014
Transgenic mouse lines subdivide external segment of the globus pallidus (GPe) neurons and reveal distinct GPe output pathways
Abstract
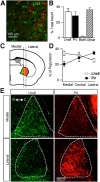
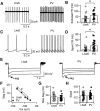
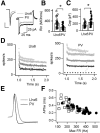
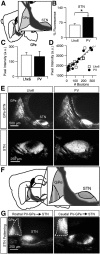
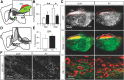
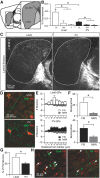
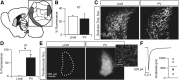
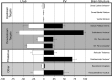
Cell-type diversity in the brain enables the assembly of complex neural circuits, whose organization and patterns of activity give rise to brain function. However, the identification of distinct neuronal populations within a given brain region is often complicated by a lack of objective criteria to distinguish one neuronal population from another. In the external segment of the globus pallidus (GPe), neuronal populations have been defined using molecular, anatomical, and electrophysiological criteria, but these classification schemes are often not generalizable across preparations and lack consistency even within the same preparation. Here, we present a novel use of existing transgenic mouse lines, Lim homeobox 6 (Lhx6)-Cre and parvalbumin (PV)-Cre, to define genetically distinct cell populations in the GPe that differ molecularly, anatomically, and electrophysiologically. Lhx6-GPe neurons, which do not express PV, are concentrated in the medial portion of the GPe. They have lower spontaneous firing rates, narrower dynamic ranges, and make stronger projections to the striatum and substantia nigra pars compacta compared with PV-GPe neurons. In contrast, PV-GPe neurons are more concentrated in the lateral portions of the GPe. They have narrower action potentials, deeper afterhyperpolarizations, and make stronger projections to the subthalamic nucleus and parafascicular nucleus of the thalamus. These electrophysiological and anatomical differences suggest that Lhx6-GPe and PV-GPe neurons participate in different circuits with the potential to contribute to different aspects of motor function and dysfunction in disease.
Keywords: basal ganglia; connectivity; globus pallidus; intrinsic excitability; transgenic mice.
Figures
References
-
- Bergman H, Wichmann T, Karmon B, DeLong MR. The primate subthalamic nucleus. II. Neuronal activity in the MPTP model of parkinsonism. J Neurophysiol. 1994;72:507–520. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
