Roles of serine and threonine residues of mumps virus P protein in viral transcription and replication
- PMID: 24501400
- PMCID: PMC3993737
- DOI: 10.1128/JVI.03673-13
Roles of serine and threonine residues of mumps virus P protein in viral transcription and replication
Abstract
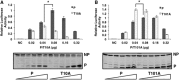
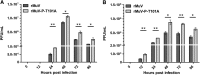
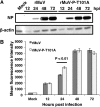
Mumps virus (MuV), a paramyxovirus containing a negative-sense nonsegmented RNA genome, is a human pathogen that causes an acute infection with symptoms ranging from parotitis to mild meningitis and severe encephalitis. Vaccination against mumps virus has been effective in reducing mumps cases. However, recently large outbreaks have occurred in vaccinated populations. There is no anti-MuV drug. Understanding replication of MuV may lead to novel antiviral strategies. MuV RNA-dependent RNA polymerase minimally consists of the phosphoprotein (P) and the large protein (L). The P protein is heavily phosphorylated. To investigate the roles of serine (S) and threonine (T) residues of P in viral RNA transcription and replication, P was subjected to mass spectrometry and mutational analysis. P, a 392-amino acid residue protein, has 64 S and T residues. We have found that mutating nine S/T residues significantly reduced and mutating residue T at 101 to A (T101A) significantly enhanced activity in a minigenome system. A recombinant virus containing the P-T101A mutation (rMuV-P-T101A) was recovered and analyzed. rMuV-P-T101A grew to higher titers and had increased protein expression at early time points. Together, these results suggest that phosphorylation of MuV-P-T101 plays a negative role in viral RNA synthesis. This is the first time that the P protein of a paramyxovirus has been systematically analyzed for S/T residues that are critical for viral RNA synthesis.
Importance: Mumps virus (MuV) is a reemerging paramyxovirus that caused large outbreaks in the United States, where vaccination coverage is very high. There is no anti-MuV drug. In this work, we have systematically analyzed roles of Ser/Thr residues of MuV P in viral RNA synthesis. We have identified S/T residues of P critical for MuV RNA synthesis and phosphorylation sites that are important for viral RNA synthesis. This work leads to a better understanding of viral RNA synthesis as well as to potential novel strategies to control mumps.
Figures


Similar articles
-
Respiratory Syncytial Virus Phosphoprotein Residue S156 Plays a Role in Regulating Genome Transcription and Replication.J Virol. 2021 Nov 23;95(24):e0120621. doi: 10.1128/JVI.01206-21. Epub 2021 Oct 6. J Virol. 2021. PMID: 34613802 Free PMC article.
-
Roles of Phosphorylation of the Nucleocapsid Protein of Mumps Virus in Regulating Viral RNA Transcription and Replication.J Virol. 2015 Jul;89(14):7338-47. doi: 10.1128/JVI.00686-15. Epub 2015 May 6. J Virol. 2015. PMID: 25948749 Free PMC article.
-
Regulation of Mumps Virus Replication and Transcription by Kinase RPS6KB1.J Virol. 2020 Jun 1;94(12):e00387-20. doi: 10.1128/JVI.00387-20. Print 2020 Jun 1. J Virol. 2020. PMID: 32295907 Free PMC article.
-
Phosphorylation of paramyxovirus phosphoprotein and its role in viral gene expression.Future Microbiol. 2010 Jan;5(1):9-13. doi: 10.2217/fmb.09.93. Future Microbiol. 2010. PMID: 20020826 Free PMC article. Review.
-
Unique Tropism and Entry Mechanism of Mumps Virus.Viruses. 2021 Sep 1;13(9):1746. doi: 10.3390/v13091746. Viruses. 2021. PMID: 34578327 Free PMC article. Review.
Cited by
-
Initiation and regulation of paramyxovirus transcription and replication.Virology. 2015 May;479-480:545-54. doi: 10.1016/j.virol.2015.01.014. Epub 2015 Feb 13. Virology. 2015. PMID: 25683441 Free PMC article. Review.
-
Oligomerization of Mumps Virus Phosphoprotein.J Virol. 2015 Nov;89(21):11002-10. doi: 10.1128/JVI.01719-15. Epub 2015 Aug 26. J Virol. 2015. PMID: 26311887 Free PMC article.
-
Advancements in the genomic feature of Newcastle disease virus and the multifaceted roles of non-structural proteins V/W in viral replication and pathogenesis.Poult Sci. 2025 Jul 3;104(10):105527. doi: 10.1016/j.psj.2025.105527. Online ahead of print. Poult Sci. 2025. PMID: 40639003 Free PMC article. Review.
-
Establishing a small animal model for evaluating protective immunity against mumps virus.PLoS One. 2017 Mar 31;12(3):e0174444. doi: 10.1371/journal.pone.0174444. eCollection 2017. PLoS One. 2017. PMID: 28362871 Free PMC article.
-
Respiratory Syncytial Virus Phosphoprotein Residue S156 Plays a Role in Regulating Genome Transcription and Replication.J Virol. 2021 Nov 23;95(24):e0120621. doi: 10.1128/JVI.01206-21. Epub 2021 Oct 6. J Virol. 2021. PMID: 34613802 Free PMC article.
References
-
- Carbone KM, Wolinsky JS. 2001. Mumps virus, p 1381–1400 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B. (ed), Fields virology, 4th ed, vol 1 Lippincott Williams and Wilkins, Philadelphia, PA
-
- MMWR. 2010. Update: mumps outbreak - New York and New Jersey, June 2009-January 2010. MMWR Morb. Mortal. Wkly. Rep. 59:125–129 - PubMed
-
- Lamb RA, Kolakofsky D. 2001. Paramyxoviridae: the viruses and their replication. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B. (ed), Fields virology, 4th ed. Lippincott, Williams and Wilkins, Philadelphia, PA
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

