Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages
- PMID: 24501410
- PMCID: PMC3993745
- DOI: 10.1128/JVI.03422-13
Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages
Abstract
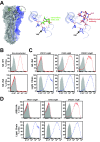
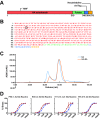
An understanding of the antigen-specific B-cell response to the influenza virus hemagglutinin (HA) is critical to the development of universal influenza vaccines, but it has not been possible to examine these cells directly because HA binds to sialic acid (SA) on most cell types. Here, we use structure-based modification of HA to isolate HA-specific B cells by flow cytometry and characterize the features of HA stem antibodies (Abs) required for their development. Incorporation of a previously described mutation (Y98F) to the receptor binding site (RBS) causes HA to bind only those B cells that express HA-specific Abs, but it does not bind nonspecifically to B cells, and this mutation has no effect on the binding of broadly neutralizing Abs to the RBS. To test the specificity of the Y98F mutation, we first demonstrated that previously described HA nanoparticles mediate hemagglutination and then determined that the Y98F mutation eliminates this activity. Cloning of immunoglobulin genes from HA-specific B cells isolated from a single human subject demonstrates that vaccination with H5N1 influenza virus can elicit B cells expressing stem monoclonal Abs (MAbs). Although these MAbs originated mostly from the IGHV1-69 germ line, a reasonable proportion derived from other genes. Analysis of stem Abs provides insight into the maturation pathways of IGVH1-69-derived stem Abs. Furthermore, this analysis shows that multiple non-IGHV1-69 stem Abs with a similar neutralizing breadth develop after vaccination in humans, suggesting that the HA stem response can be elicited in individuals with non-stem-reactive IGHV1-69 alleles.
Importance: Universal influenza vaccines would improve immune protection against infection and facilitate vaccine manufacturing and distribution. Flu vaccines stimulate B cells in the blood to produce antibodies that neutralize the virus. These antibodies target a protein on the surface of the virus called HA. Flu vaccines must be reformulated annually, because these antibodies are mostly specific to the viral strains used in the vaccine. But humans can produce broadly neutralizing antibodies. We sought to isolate B cells whose genes encode influenza virus antibodies from a patient vaccinated for avian influenza. To do so, we modified HA so it would bind only the desired cells. Sequencing the antibody genes of cells marked by this probe proved that the patient produced broadly neutralizing antibodies in response to the vaccine. Many sequences obtained had not been observed before. There are more ways to generate broadly neutralizing antibodies for influenza virus than previously thought.
Figures


References
-
- Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, Marasco WA. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16:265–273. 10.1038/nsmb.1566 - DOI - PMC - PubMed
-
- Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, Lanzavecchia A. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856. 10.1126/science.1205669 - DOI - PubMed
-
- Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, Vogels R, Brakenhoff JP, Kompier R, Koldijk MH, Cornelissen LA, Poon LL, Peiris M, Koudstaal W, Wilson IA, Goudsmit J. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333:843–850. 10.1126/science.1204839 - DOI - PMC - PubMed
-
- Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, Kepler TB, Liao HX, Harrison SC. 2011. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. U. S. A. 108:14216–14221. 10.1073/pnas.1111497108 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

