Flexibility in surface-exposed loops in a virus capsid mediates escape from antibody neutralization
- PMID: 24501415
- PMCID: PMC3993751
- DOI: 10.1128/JVI.03685-13
Flexibility in surface-exposed loops in a virus capsid mediates escape from antibody neutralization
Abstract
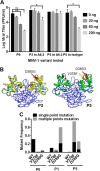
New human norovirus strains emerge every 2 to 3 years, partly due to mutations in the viral capsid that allow escape from antibody neutralization and herd immunity. To understand how noroviruses evolve antibody resistance, we investigated the structural basis for the escape of murine norovirus (MNV) from antibody neutralization. To identify specific residues in the MNV-1 protruding (P) domain of the capsid that play a role in escape from the neutralizing monoclonal antibody (MAb) A6.2, 22 recombinant MNVs were generated with amino acid substitutions in the A'B' and E'F' loops. Six mutations in the E'F' loop (V378F, A382K, A382P, A382R, D385G, and L386F) mediated escape from MAb A6.2 neutralization. To elucidate underlying structural mechanisms for these results, the atomic structure of the A6.2 Fab was determined and fitted into the previously generated pseudoatomic model of the A6.2 Fab/MNV-1 virion complex. Previously, two distinct conformations, A and B, of the atomic structures of the MNV-1 P domain were identified due to flexibility in the two P domain loops. A superior stereochemical fit of the A6.2 Fab to the A conformation of the MNV P domain was observed. Structural analysis of our observed escape mutants indicates changes toward the less-preferred B conformation of the P domain. The shift in the structural equilibrium of the P domain toward the conformation with poor structural complementarity to the antibody strongly supports a unique mechanism for antibody escape that occurs via antigen flexibility instead of direct antibody-antigen binding.
Importance: Human noroviruses cause the majority of all nonbacterial gastroenteritis worldwide. New epidemic strains arise in part by mutations in the viral capsid leading to escape from antibody neutralization. Herein, we identify a series of point mutations in a norovirus capsid that mediate escape from antibody neutralization and determine the structure of a neutralizing antibody. Fitting of the antibody structure into the virion/antibody complex identifies two conformations of the antibody binding domain of the viral capsid: one with a superior fit and the other with an inferior fit to the antibody. These data suggest a unique mode of antibody neutralization. In contrast to other viruses that largely escape antibody neutralization through direct disruption of the antibody-virus interface, we identify mutations that acted indirectly by limiting the conformation of the antibody binding loop in the viral capsid and drive the antibody binding domain into the conformation unable to be bound by the antibody.
Figures




References
-
- Debbink K, Lindesmith LC, Donaldson EF, Costantini V, Beltramello M, Corti D, Swanstrom J, Lanzavecchia A, Vinje J, Baric RS. 2013. Emergence of new pandemic GII.4 Sydney norovirus strain correlates with escape from herd immunity. J. Infect. Dis. 208:1877–1887. 10.1093/infdis/jit370 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

