Differential escape patterns within the dominant HLA-B*57:03-restricted HIV Gag epitope reflect distinct clade-specific functional constraints
- PMID: 24501417
- PMCID: PMC3993828
- DOI: 10.1128/JVI.03303-13
Differential escape patterns within the dominant HLA-B*57:03-restricted HIV Gag epitope reflect distinct clade-specific functional constraints
Abstract
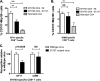
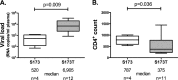
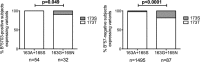
HLA-B*57:01 and HLA-B*57:03, the most prevalent HLA-B*57 subtypes in Caucasian and African populations, respectively, are the HLA alleles most protective against HIV disease progression. Understanding the mechanisms underlying this immune control is of critical importance, yet they remain unclear. Unexplained differences are observed in the impact of the dominant cytotoxic T lymphocyte (CTL) response restricted by HLA-B*57:01 and HLA-B*57:03 in chronic infection on the Gag epitope KAFSPEVIPMF (KF11; Gag 162 to 172). We previously showed that the HLA-B*57:03-KF11 response is associated with a >1-log-lower viral setpoint in C clade virus infection and that this response selects escape mutants within the epitope. We first examined the relationship of KF11 responses in B clade virus-infected subjects with HLA-B*57:01 to immune control and observed that a detectable KF11 response was associated with a >1-log-higher viral load (P = 0.02). No evidence of HLA-B*57:01-KF11-associated selection pressure was identified in previous comprehensive analyses of >1,800 B clade virus-infected subjects. We then studied a B clade virus-infected cohort in Barbados, where HLA-B*57:03 is highly prevalent. In contrast to findings for B clade virus-infected subjects expressing HLA-B*57:01, we observed strong selection pressure driven by the HLA-B*57:03-KF11 response for the escape mutation S173T. This mutation reduces recognition of virus-infected cells by HLA-B*57:03-KF11 CTLs and is associated with a >1-log increase in viral load in HLA-B*57:03-positive subjects (P = 0.009). We demonstrate functional constraints imposed by HIV clade relating to the residue at Gag 173 that explain the differential clade-specific escape patterns in HLA-B*57:03 subjects. Further studies are needed to evaluate the role of the KF11 response in HLA-B*57:01-associated HIV disease protection.
Importance: HLA-B*57 is the HLA class I molecule that affords the greatest protection against disease progression in HIV infection. Understanding the key mechanism(s) underlying immunosuppression of HIV is of importance in guiding therapeutic and vaccine-related approaches to improve the levels of HIV control occurring in nature. Numerous mechanisms have been proposed to explain the HLA associations with differential HIV disease outcome, but no consensus exists. These studies focus on two subtypes of HLA-B*57 prevalent in Caucasian and African populations, HLA-B*57:01 and HLA-B*57:03, respectively. These alleles appear equally protective against HIV disease progression. The CTL epitopes presented are in many cases identical, and the dominant response in chronic infection in each case is to the Gag epitope KF11. However, there the similarity ends. This study sought to better understand the reasons for these differences and what they teach us about which immune responses contribute to immune control of HIV infection.
Figures



References
-
- Kaslow RA, Carrington M, Apple R, Park L, Munoz A, Saah AJ, Goedert JJ, Winkler C, O'Brien SJ, Rinaldo C, Detels R, Blattner W, Phair J, Erlich H, Mann DL. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405–411. 10.1038/nm0496-405 - DOI - PubMed
-
- Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, Martino L, Hallahan CW, Selig SM, Schwartz D, Sullivan J, Connors M. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. U. S. A. 97:2709–2714. 10.1073/pnas.050567397 - DOI - PMC - PubMed
-
- Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, Zimbwa P, Moore S, Allen T, Brander C, Addo MM, Altfeld M, James I, Mallal S, Bunce M, Barber LD, Szinger J, Day C, Klenerman P, Mullins J, Korber B, Coovadia HM, Walker BD, Goulder PJ. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769–775. 10.1038/nature03113 - DOI - PubMed
-
- Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, Kadie CM, Carlson JM, Heckerman D, Graham RR, Plenge RM, Deeks SG, Gianniny L, Crawford G, Sullivan J, Gonzalez E, Davies L, Camargo A, Moore JM, Beattie N, Gupta S, Crenshaw A, Burtt NP, Guiducci C, Gupta N, Gao X, Qi Y, Yuki Y, Piechocka-Trocha A, Cutrell E, Rosenberg R, Moss KL, Lemay P, O'Leary J, Schaefer T, Verma P, Toth I, Block B, Baker B, Rothchild A, Lian J, Proudfoot J, Alvino DM, Vine S, Addo MM, Allen TM, et al. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557. 10.1126/science.1195271 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

