Neural cell alignment by patterning gradients of the extracellular matrix protein laminin
- PMID: 24501672
- PMCID: PMC3886309
- DOI: 10.1098/rsfs.2013.0041
Neural cell alignment by patterning gradients of the extracellular matrix protein laminin
Abstract
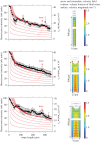
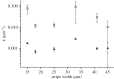
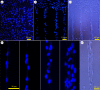
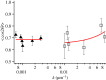
Anisotropic orientation and accurate positioning of neural cells is achieved by patterning stripes of the extracellular matrix protein laminin on the surface of polystyrene tissue culture dishes by micromoulding in capillaries (MIMICs). Laminin concentration decreases from the entrance of the channels in contact with the reservoir towards the end. Immunofluorescence analysis of laminin shows a decreasing gradient of concentration along the longitudinal direction of the stripes. The explanation is the superposition of diffusion and convection of the solute, the former dominating at length scales near the entrance (characteristic length around 50 μm), the latter further away (length scale in excess of 900 μm). These length scales are independent of the channel width explored from about 15 to 45 μm. Neural cells are randomly seeded and selectively adhere to the pattern, leaving the unpatterned areas depleted even upon 6 days of incubation. Cell alignment was assessed by the orientation of the long axis of the 4',6-diamidino-2-phenylindole-stained nuclei. Samples on patterned the laminin area exhibit a large orientational order parameter. As control, cells on the unpatterned laminin film exhibit no preferential orientation. This implies that the anisotropy of laminin stripes is an effective chemical stimulus for cell recruiting and alignment.
Keywords: laminin; neural cells; patterning; protein gradients; soft lithography.
Figures


Similar articles
-
Immobilized laminin concentration gradients on electrospun fiber scaffolds for controlled neurite outgrowth.Biointerphases. 2014 Mar;9(1):011003. doi: 10.1116/1.4857295. Biointerphases. 2014. PMID: 24739010
-
Guided cell adhesion, orientation, morphology and differentiation on silicon substrates photolithographically micropatterned with a cell-repellent cross-linked poly(vinyl alcohol) film.Biomed Mater. 2018 Oct 26;14(1):014101. doi: 10.1088/1748-605X/aae7ba. Biomed Mater. 2018. PMID: 30362459
-
Astrocyte spreading and migration on aggrecan-laminin dot gradients.Biointerphases. 2017 Sep 11;13(1):01A401. doi: 10.1116/1.5001675. Biointerphases. 2017. PMID: 28893070 Free PMC article.
-
How the zebra got its stripes: Curvature-dependent diffusion orients Turing patterns on three-dimensional surfaces.Phys Rev E. 2024 Sep;110(3-1):034402. doi: 10.1103/PhysRevE.110.034402. Phys Rev E. 2024. PMID: 39425380
-
Imaging via widefield surface plasmon resonance microscope for studying bone cell interactions with micropatterned ECM proteins.J Microsc. 2011 Mar;241(3):282-90. doi: 10.1111/j.1365-2818.2010.03430.x. J Microsc. 2011. PMID: 21118224
Cited by
-
Coextruded, aligned, and gradient-modified poly(ε-caprolactone) fibers as platforms for neural growth.Biomacromolecules. 2015 Mar 9;16(3):860-7. doi: 10.1021/bm501767x. Epub 2015 Feb 26. Biomacromolecules. 2015. PMID: 25715836 Free PMC article.
-
Automated Robotic Dispensing Technique for Surface Guidance and Bioprinting of Cells.J Vis Exp. 2016 Nov 18;(117):54604. doi: 10.3791/54604. J Vis Exp. 2016. PMID: 27911405 Free PMC article.
-
Enzyme-Mediated Conjugation of Peptides to Silk Fibroin for Facile Hydrogel Functionalization.Ann Biomed Eng. 2020 Jul;48(7):1905-1915. doi: 10.1007/s10439-020-02503-2. Epub 2020 Apr 20. Ann Biomed Eng. 2020. PMID: 32314301 Free PMC article.
-
Functionalized α-Helical Peptide Hydrogels for Neural Tissue Engineering.ACS Biomater Sci Eng. 2015 Jun 8;1(6):431-439. doi: 10.1021/acsbiomaterials.5b00051. Epub 2015 Apr 28. ACS Biomater Sci Eng. 2015. PMID: 26240838 Free PMC article.
-
PEDOT: PSS promotes neurogenic commitment of neural crest-derived stem cells.Front Physiol. 2022 Aug 17;13:930804. doi: 10.3389/fphys.2022.930804. eCollection 2022. Front Physiol. 2022. PMID: 36060701 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

