Biochemical and functional analyses of gp130 mutants unveil JAK1 as a novel therapeutic target in human inflammatory hepatocellular adenoma
- PMID: 24501689
- PMCID: PMC3913689
- DOI: 10.4161/onci.27090
Biochemical and functional analyses of gp130 mutants unveil JAK1 as a novel therapeutic target in human inflammatory hepatocellular adenoma
Abstract
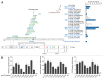
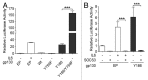
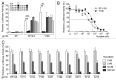
Inflammatory hepatocellular adenomas (IHCAs) are benign liver lesions that can be characterized histologically by the presence of an inflammatory infiltrate and at the molecular level by the overexpression of acute phase inflammatory response genes. Recurrent somatic mutations of the interleukin-6 (IL-6) signal transducer (IL6ST) locus, encoding the critical component of the IL-6 signal transduction machinery gp130, are present in 60% of IHCAs and in a subset (2%) of hepatocellular carcinoma (HCCs). By screening of 256 human hepatic adenoma specimens (the largest genetic analysis of IL6ST performed to date in this setting), we identified 24 distinct somatic IL6ST mutations among 66 mutant adenomas. The functional analysis of nine different gp130 mutants expressed in hepatic cancer cell lines consistently revealed the constitutive and IL-6-independent activation of the JAK/STAT signaling pathway. We further demonstrated that the signaling activity of mutant gp130 in IHCA remains responsive to suppressor of cytokine signaling 3 (SOCS3), a physiological gp130 inhibitor. Specifically, cells expressing a double mutant variant of gp130 with a disrupted SOCS3-binding site at residue 759 (Y186/Y759F) displayed a hyperactivation of signal transducer and activator of transcription 3 (STAT3) as compared with cells expressing the endogenous IHCA-associated Y186 gp130 mutant. Notably, we identified that constitutive signaling via gp130 in IHCA requires the Janus kinase family member JAK1, but not JAK2 or tyrosine kinase 2. In support of this notion, AG490, a tyrosine kinase inhibitor that selectively blocks JAK2, had no effect on gp130 activity. In stark contrast, we showed that ruxolitinib, a JAK1/JAK2-selective tyrosine kinase inhibitor used to treat patients with myelofibrosis, dramatically impaired JAK1-STAT signaling downstream of all IHCA-associated gp130 mutants. In conclusion, our findings provide a rationale for the use of JAK1 inhibitors for the treatment of HCAs expressing mutant gp130 as well as a subset of HCCs that bear similar mutations.
Keywords: IL6ST; JAK1; STAT3; ruxolitinib; targeted therapy.
Figures

Similar articles
-
Mutations leading to constitutive active gp130/JAK1/STAT3 pathway.Cytokine Growth Factor Rev. 2015 Oct;26(5):499-506. doi: 10.1016/j.cytogfr.2015.07.010. Epub 2015 Jul 3. Cytokine Growth Factor Rev. 2015. PMID: 26188635 Review.
-
Somatic mutations activating STAT3 in human inflammatory hepatocellular adenomas.J Exp Med. 2011 Jul 4;208(7):1359-66. doi: 10.1084/jem.20110283. Epub 2011 Jun 20. J Exp Med. 2011. PMID: 21690253 Free PMC article.
-
Activation of the protein tyrosine phosphatase SHP2 via the interleukin-6 signal transducing receptor protein gp130 requires tyrosine kinase Jak1 and limits acute-phase protein expression.Biochem J. 1998 Nov 1;335 ( Pt 3)(Pt 3):557-65. doi: 10.1042/bj3350557. Biochem J. 1998. PMID: 9794795 Free PMC article.
-
Recurrent chromosomal rearrangements of ROS1, FRK and IL6 activating JAK/STAT pathway in inflammatory hepatocellular adenomas.Gut. 2020 Sep;69(9):1667-1676. doi: 10.1136/gutjnl-2019-319790. Epub 2020 Jan 6. Gut. 2020. PMID: 31907296
-
Inhibition of IL-6 family cytokines by SOCS3.Semin Immunol. 2014 Feb;26(1):13-9. doi: 10.1016/j.smim.2013.12.004. Epub 2014 Jan 10. Semin Immunol. 2014. PMID: 24418198 Free PMC article. Review.
Cited by
-
Management of hepatic adenomatosis.Curr Gastroenterol Rep. 2015 Mar;17(3):12. doi: 10.1007/s11894-015-0434-4. Curr Gastroenterol Rep. 2015. PMID: 25740249 Review.
-
Intracellular signaling prevents effective blockade of oncogenic gp130 mutants by neutralizing antibodies.Cell Commun Signal. 2014 Mar 10;12:14. doi: 10.1186/1478-811X-12-14. Cell Commun Signal. 2014. PMID: 24612692 Free PMC article.
-
Molecular classification of hepatocellular adenomas: impact on clinical practice.Hepat Oncol. 2018 Apr 9;5(1):HEP04. doi: 10.2217/hep-2017-0023. eCollection 2018 Jan. Hepat Oncol. 2018. PMID: 30302195 Free PMC article. Review.
-
Inflammatory hepatocellular adenomas developed in the setting of chronic liver disease and cirrhosis.Mod Pathol. 2016 Jan;29(1):43-50. doi: 10.1038/modpathol.2015.119. Epub 2015 Oct 30. Mod Pathol. 2016. PMID: 26516697
-
A cell-autonomous tumour suppressor role of RAF1 in hepatocarcinogenesis.Nat Commun. 2016 Dec 21;7:13781. doi: 10.1038/ncomms13781. Nat Commun. 2016. PMID: 28000790 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
