Transcriptomic response to prolonged ethanol production in the cyanobacterium Synechocystis sp. PCC6803
- PMID: 24502290
- PMCID: PMC3925133
- DOI: 10.1186/1754-6834-7-21
Transcriptomic response to prolonged ethanol production in the cyanobacterium Synechocystis sp. PCC6803
Abstract
Background: The production of biofuels in photosynthetic microalgae and cyanobacteria is a promising alternative to the generation of fuels from fossil resources. To be economically competitive, producer strains need to be established that synthesize the targeted product at high yield and over a long time. Engineering cyanobacteria into forced fuel producers should considerably interfere with overall cell homeostasis, which in turn might counteract productivity and sustainability of the process. Therefore, in-depth characterization of the cellular response upon long-term production is of high interest for the targeted improvement of a desired strain.
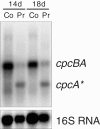
Results: The transcriptome-wide response to continuous ethanol production was examined in Synechocystis sp. PCC6803 using high resolution microarrays. In two independent experiments, ethanol production rates of 0.0338% (v/v) ethanol d-1 and 0.0303% (v/v) ethanol d-1 were obtained over 18 consecutive days, measuring two sets of biological triplicates in fully automated photobioreactors. Ethanol production caused a significant (~40%) delay in biomass accumulation, the development of a bleaching phenotype and a down-regulation of light harvesting capacity. However, microarray analyses performed at day 4, 7, 11 and 18 of the experiment revealed only three mRNAs with a strongly modified accumulation level throughout the course of the experiment. In addition to the overexpressed adhA (slr1192) gene, this was an approximately 4 fold reduction in cpcB (sll1577) and 3 to 6 fold increase in rps8 (sll1809) mRNA levels. Much weaker modifications of expression level or modifications restricted to day 18 of the experiment were observed for genes involved in carbon assimilation (Ribulose bisphosphate carboxylase and Glutamate decarboxylase). Molecular analysis of the reduced cpcB levels revealed a post-transcriptional processing of the cpcBA operon mRNA leaving a truncated mRNA cpcA* likely not competent for translation. Moreover, western blots and zinc-enhanced bilin fluorescence blots confirmed a severe reduction in the amounts of both phycocyanin subunits, explaining the cause of the bleaching phenotype.
Conclusions: Changes in gene expression upon induction of long-term ethanol production in Synechocystis sp. PCC6803 are highly specific. In particular, we did not observe a comprehensive stress response as might have been expected.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

