The HicA toxin from Burkholderia pseudomallei has a role in persister cell formation
- PMID: 24502667
- PMCID: PMC3969222
- DOI: 10.1042/BJ20140073
The HicA toxin from Burkholderia pseudomallei has a role in persister cell formation
Abstract
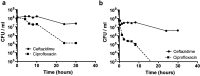
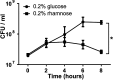
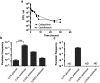
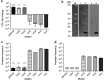
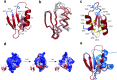
TA (toxin-antitoxin) systems are widely distributed amongst bacteria and are associated with the formation of antibiotic tolerant (persister) cells that may have involvement in chronic and recurrent disease. We show that overexpression of the Burkholderia pseudomallei HicA toxin causes growth arrest and increases the number of persister cells tolerant to ciprofloxacin or ceftazidime. Furthermore, our data show that persistence towards ciprofloxacin or ceftazidime can be differentially modulated depending on the level of induction of HicA expression. Deleting the hicAB locus from B. pseudomallei K96243 significantly reduced persister cell frequencies following exposure to ciprofloxacin, but not ceftazidime. The structure of HicA(H24A) was solved by NMR and forms a dsRBD-like (dsRNA-binding domain-like) fold, composed of a triple-stranded β-sheet, with two helices packed against one face. The surface of the protein is highly positively charged indicative of an RNA-binding protein and His24 and Gly22 were functionality important residues. This is the first study demonstrating a role for the HicAB system in bacterial persistence and the first structure of a HicA protein that has been experimentally characterized.
Figures

References
-
- Wiersinga W., van der Poll T., White N., Day N., Peacock S. J. Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat. Rev. Microbiol. 2006;4:273–282. - PubMed
-
- Wiersinga W. J., Currie B. J., Peacock S. J. Melioidosis. N. Engl. J. Med. 2012;367:1035–1044. - PubMed
-
- Limmathurotsakul D., Peacock S. J. Melioidosis: a clinical overview. Br. Med. Bull. 2011;99:125–139. - PubMed
-
- Currie B., Fisher D., Anstey N., Jacups S. Melioidosis: acute and chronic disease, relapse and re-activation. Trans. R. Soc. Trop. Med. Hyg. 2000;94:301–304. - PubMed
-
- Lazar Adler N. R., Govan B., Cullinane M., Harper M., Adler B., Boyce J. D. The molecular and cellular basis of pathogenesis in melioidosis: how does Burkholderia pseudomallei cause disease? FEMS Microbiol. Rev. 2009;33:1079–1099. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

