Direct and indirect targets of the E2A-PBX1 leukemia-specific fusion protein
- PMID: 24503810
- PMCID: PMC3913655
- DOI: 10.1371/journal.pone.0087602
Direct and indirect targets of the E2A-PBX1 leukemia-specific fusion protein
Abstract
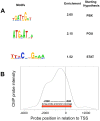
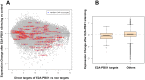
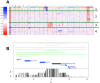
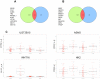
E2A-PBX1 is expressed as a result of the t(1;19) chromosomal translocation in nearly 5% of cases of childhood acute lymphoblastic leukemia. The E2A-PBX1 chimeric transcription factor contains the N-terminal transactivation domain of E2A (TCF3) fused to the C-terminal DNA-binding homeodomain of PBX1. While there is no doubt of its oncogenic potential, the mechanisms of E2A-PBX1-mediated pre-B cell transformation and the nature of direct E2A-PBX1 target genes and pathways remain largely unknown. Herein we used chromatin immunoprecipitation assays (ChIP-chip) to identify direct targets of E2A-PBX1, and we used gene expression arrays of siRNA E2A-PBX1-silenced cells to evaluate changes in expression induced by the fusion protein. Combined ChIP-chip and expression data analysis gave rise to direct and functional targets of E2A-PBX1. Further we observe that the set of ChIP-chip identified E2A-PBX1 targets show a collective down-regulation trend in the E2A-PBX1 silenced samples compared to controls suggesting an activating role of this fusion transcription factor. Our data suggest that the expression of the E2A-PBX1 fusion gene interferes with key regulatory pathways and functions of hematopoietic biology. Among these are members of the WNT and apoptosis/cell cycle control pathways, and thus may comprise an essential driving force for the propagation and maintenance of the leukemic phenotype. These findings may also provide evidence of potentially attractive therapeutic targets.
Conflict of interest statement
Figures

Similar articles
-
Mediator subunit MED1 is required for E2A-PBX1-mediated oncogenic transcription and leukemic cell growth.Proc Natl Acad Sci U S A. 2021 Feb 9;118(6):e1922864118. doi: 10.1073/pnas.1922864118. Proc Natl Acad Sci U S A. 2021. PMID: 33542097 Free PMC article.
-
E2A-PBX1 functions as a coactivator for RUNX1 in acute lymphoblastic leukemia.Blood. 2020 Jul 2;136(1):11-23. doi: 10.1182/blood.2019003312. Blood. 2020. PMID: 32276273 Free PMC article.
-
The highest affinity DNA element bound by Pbx complexes in t(1;19) leukemic cells fails to mediate cooperative DNA-binding or cooperative transactivation by E2a-Pbx1 and class I Hox proteins - evidence for selective targetting of E2a-Pbx1 to a subset of Pbx-recognition elements.Oncogene. 1997 May 29;14(21):2521-31. doi: 10.1038/sj.onc.1201097. Oncogene. 1997. PMID: 9191052
-
The advances of E2A-PBX1 fusion in B-cell acute lymphoblastic Leukaemia.Ann Hematol. 2024 Sep;103(9):3385-3398. doi: 10.1007/s00277-023-05595-7. Epub 2023 Dec 27. Ann Hematol. 2024. PMID: 38148344 Review.
-
The role of E2A-PBX1 in leukemogenesis.Oncogene. 2001 Sep 10;20(40):5708-17. doi: 10.1038/sj.onc.1204592. Oncogene. 2001. PMID: 11607820 Review. No abstract available.
Cited by
-
E2A-PBX1 exhibited a promising prognosis in pediatric acute lymphoblastic leukemia treated with the CCLG-ALL2008 protocol.Onco Targets Ther. 2016 Nov 22;9:7219-7225. doi: 10.2147/OTT.S115257. eCollection 2016. Onco Targets Ther. 2016. PMID: 27920559 Free PMC article.
-
PKCζ and PKMζ are overexpressed in TCF3-rearranged paediatric acute lymphoblastic leukaemia and are associated with increased thiopurine sensitivity.Leukemia. 2015 Feb;29(2):304-11. doi: 10.1038/leu.2014.210. Epub 2014 Jul 3. Leukemia. 2015. PMID: 24990612 Free PMC article.
-
High frequency and poor prognosis of late childhood BCR-ABL-positive and MLL-AF4-positive ALL define the need for advanced molecular diagnostics and improved therapeutic strategies in pediatric B-ALL in Pakistan.Mol Diagn Ther. 2015 Oct;19(5):277-87. doi: 10.1007/s40291-015-0149-0. Mol Diagn Ther. 2015. PMID: 26266519
-
Targeting TNF/IL-17/MAPK pathway in hE2A-PBX1 leukemia: effects of OUL35, KJ-Pyr-9, and CID44216842.Haematologica. 2024 Jul 1;109(7):2092-2110. doi: 10.3324/haematol.2023.283647. Haematologica. 2024. PMID: 38385270 Free PMC article.
-
Comprehensive summary: the role of PBX1 in development and cancers.Front Cell Dev Biol. 2024 Jul 26;12:1442052. doi: 10.3389/fcell.2024.1442052. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39129784 Free PMC article. Review.
References
-
- Aspland SE, Bendall HH, Murre C (2001) The role of E2A-PBX1 in leukemogenesis. Oncogene 20: 5708–5717. - PubMed
-
- Carroll WL, Bhojwani D, Min DJ, Moskowitz N, Raetz EA (2006) Childhood acute lymphoblastic leukemia in the age of genomics. Pediatr Blood Cancer 46: 570–578. - PubMed
-
- Harrison CJ (2001) The detection and significance of chromosomal abnormalities in childhood acute lymphoblastic leukaemia. Blood Rev 15: 49–59. - PubMed
-
- Bourette RP, Grasset MF, Mouchiroud G (2007) E2a/Pbx1 oncogene inhibits terminal differentiation but not myeloid potential of pro-T cells. Oncogene 26: 234–247. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

