Molecular profiling of blastic plasmacytoid dendritic cell neoplasm reveals a unique pattern and suggests selective sensitivity to NF-kB pathway inhibition
- PMID: 24504027
- PMCID: PMC4294271
- DOI: 10.1038/leu.2014.64
Molecular profiling of blastic plasmacytoid dendritic cell neoplasm reveals a unique pattern and suggests selective sensitivity to NF-kB pathway inhibition
Abstract
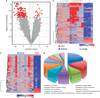
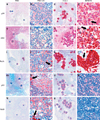
Blastic plasmacytoid dendritic cell neoplasm (BPDCN) is a rare disease of controversial origin recently recognized as a neoplasm deriving from plasmacytoid dendritic cells (pDCs). Nevertheless, it remains an orphan tumor with obscure biology and dismal prognosis. To better understand the pathobiology of BPDCN and discover new targets for effective therapies, the gene expression profile (GEP) of 25 BPDCN samples was analyzed and compared with that of pDCs, their postulated normal counterpart. Validation was performed by immunohistochemistry (IHC), whereas functional experiments were carried out ex vivo. For the first time at the molecular level, we definitely recognized the cellular derivation of BPDCN that proved to originate from the myeloid lineage and in particular, from resting pDCs. Furthermore, thanks to an integrated bioinformatic approach we discovered aberrant activation of the NF-kB pathway and suggested it as a novel therapeutic target. We tested the efficacy of anti-NF-kB-treatment on the BPDCN cell line CAL-1, and successfully demonstrated by GEP and IHC the molecular shutoff of the NF-kB pathway. In conclusion, we identified a molecular signature representative of the transcriptional abnormalities of BPDCN and developed a cellular model proposing a novel therapeutic approach in the setting of this otherwise incurable disease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Facchetti FJD, Petrella T. Blastic plasmacytoid dendritic cell neoplasms. In: Swerdlow SH, Campo E, Harris NL, et al., editors. WHO Classification of Tumors of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC; 2008. pp. 145–147.
-
- Cella M, Facchetti F, Lanzavecchia A, Colonna M. Plasmacytoid dendritic cells activated by influenza virus and CD40L drive a potent TH1 polarization. Nat Immunol. 2000;1:305–310. - PubMed
-
- Soumelis V, Liu YJ. From plasmacytoid to dendritic cell: morphological and functional switches during plasmacytoid pre-dendritic cell differentiation. Eur J Immunol. 2006;36:2286–2292. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

